Clustering in Two-mode Networks
A subject that has long received attention in both theoretical and empirical research is nodes’ tendency to cluster together. Evidence suggests that in most real-world networks, and especially in social networks, nodes cluster into densely connected groups (Holland and Leinhardt, 1970; Opsahl and Panzarasa, 2009). A number of measures have been developed for testing this tendency. Specifically, the global clustering coefficient assesses the overall level of clustering in a network (Luce and Perry, 1949), and the local clustering coefficient assesses the clustering in a single node’s immediate network (i.e., the node and its contacts; Watts and Strogatz, 1998). Formally, the global coefficient is the fraction of triplets or 2-paths (i.e., three nodes connected by two ties) that are closed by the presence of a tie between the first and the third node, and the local clustering coefficient is the fraction of 2-paths centered on the focal node that are closed by the presence of a tie between the first and the third node. For a review, see Opsahl and Panzarasa (2009) or the Clustering in Weighted Networks-page.
Applying the two clustering coefficients directly to a two-mode network is senseless as two-mode networks are bipartite and, thus, the contacts of a node cannot be connected to each other by construction and no triangles can exist (Borgatti and Everett, 1997). When one-mode measures cannot directly be applied to two-mode networks, there are two options: (1) transform two-mode networks into one-mode networks so that the one-mode measure can be used and (2) use the specially designed measures for two-mode networks.
Why not transform two-mode networks into one-mode networks when measuring clustering
A number of issues are present when measuring clustering in transformed two-mode network. Transforming a two-mode network to a one-mode network is often done using a method known as projection. This method operates by selecting the primary nodes and linking two nodes if they have at least one common secondary node. Projected two-mode network tends to have more and larger fully-connected cliques than prototypical one-mode networks (Wasserman and Faust, 1994). This has direct implications for network measures, especially those based on triangles including the structural holes measures (Burt, 1992, 2005) and the clustering coefficients (Opsahl and Panzarasa, 2009). To exemplify the cliques, and the many closed triplets, produced when projecting a two-mode network, this diagram shows the main component of the interpersonal network among Norwegian directors (Seierstad and Opsahl, 2011).

The Norwegian interlocking directorate on August 1, 2009. The network structure among directors (circles) who form part of the largest group of interconnected directors. Two directors are connected if they are members of the same board. The solid circles refer to women, whereas the hollow circles refer to men. Available through boardsandgender.com.
To further explore whether projected networks contain many more triangles than prototypical networks with a similar tendency for triadic closure, I randomized the two-mode structure of a scientific collaboration network (Newman, 2001) while maintaining the degree distributions (i.e., randomly assigning the ties in the two-mode network while keeping each author’s number of co-authored papers, and each paper’s number of authors; see Random Two-mode Networks) before projecting it onto a one-mode network and calculating the global clustering coefficient. By using this randomization procedure, the triangles formed due to the two-mode structure are maintained in the one-mode projection. Across 1,000 projected random two-mode networks, the average global clustering coefficient was 0.1236, which is over 350 times larger than the coefficient in corresponding one-mode classical random networks. As such, the default benchmark value of the clustering coefficient is not appropriate when projecting a two-mode network.

Based on random versions of Newman's (2001) scientific collaboration network. The values can be fitted with 1.02degree^{-0.93} with an R² of 0.9881.
Clustering coefficients for two-mode networks: Reinforcement
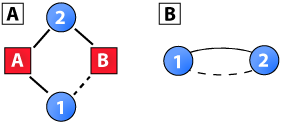 To overcome this bias, a number of clustering coefficients for two-mode networks have been proposed in the literature (Latapy et al., 2008; Lind et al., 2005; Opsahl, 2012; Robins and Alexander, 2004; Zhang et al., 2008). The first set of clustering coefficients for two-mode networks are based on 4-cycles, which is the smallest possible cycle in two-mode networks. For example, Robins and Alexander (2004) defined a coefficient as the ratio between the number of 4-cycles and the number of 3-paths. This measure is illustrated in Panel A of the diagram to the right. The solid lines represent a 3-path and the count of these in a network would be the denominator. If the dashed line was present, the 3-path would be closed and part of a 4-cycle. The count of these would be the numerator. Formally, this coefficient is:
To overcome this bias, a number of clustering coefficients for two-mode networks have been proposed in the literature (Latapy et al., 2008; Lind et al., 2005; Opsahl, 2012; Robins and Alexander, 2004; Zhang et al., 2008). The first set of clustering coefficients for two-mode networks are based on 4-cycles, which is the smallest possible cycle in two-mode networks. For example, Robins and Alexander (2004) defined a coefficient as the ratio between the number of 4-cycles and the number of 3-paths. This measure is illustrated in Panel A of the diagram to the right. The solid lines represent a 3-path and the count of these in a network would be the denominator. If the dashed line was present, the 3-path would be closed and part of a 4-cycle. The count of these would be the numerator. Formally, this coefficient is:
This measure departs from the concept of triadic closure as a 3-path would simply be, in the case of scientific collaboration networks, the number of papers that an author’s collaborators have co-authored, and a 4-cycle would be two authors collaborating twice (see Panel B of the diagram). Although this could be viewed as a form of clustering, it would not be triadic closure as it includes only two individuals. In fact, it could be considered a measure of reinforcement between two individuals rather than clustering of a group of individuals. From an evolving network perspective (e.g., in ERG models and SIENA; Peng et al., 2009; Snijders et al., 2011), it could be conceptualized as the adaptation or agreement that leads a node to connect to the same group as others.
Clustering coefficients for two-mode networks: Global coefficient

(a) A two-mode network where the shape and color of nodes represent the node set to which a node belongs, and (b) the one-mode projection of the round blue nodes in the two-mode network in panel A. The round blue nodes are the primary nodes in this projection. (c) A weighted two-mode network with a similar topology as the binary two-mode network in panel A.
where 4-paths that are closed by being part of at least one 6-cycle (i.e., a loop of six ties with five nodes).
The coefficient has a number of properties. First, it varies between 0 and 1 as the numerator and denominator are positive numbers, and the numerator is a subset of the denominator. Second, in a fully-connected network, it is equal to 1 as all 4-paths are closed. Third, in classical random two-mode networks (i.e., with a set number of nodes and density), the expected value is , where
is the number of secondary nodes and d is the density of the two-mode network (Borgatti and Everett, 1997).
The one-mode global clustering coefficient has been extended to weighted networks by assigning a value to each triplet or 2-path (Opsahl and Panzarasa, 2009). This value is based on the weights of the two ties that compose the 2-path (e.g., the arithmetic mean, geometric mean, maximum, or minimum of the tie weights). The coefficient incorporates these values by being defined as the total value of closed 2-paths over the total value of all 2-paths. In addition to having the same parameters as the original coefficient, this coefficient produces the same outcome as the original one if all 2-paths have the same value (e.g., 1 in a binary network) or if the weights are randomly reshuffled in the network. In a similar spirit, the global two-mode coefficient can be generalized to weighted two-mode networks, such as those created from online forums where the weights are the number of messages or characters posted to a thread. Specifically, a 4-path-value could be defined instead of a 2-path value. This value should be constructed based on the four tie weights, and could be defined using the same four methods as the one-mode weighted clustering coefficient. For example, the 4-path from node 1 to node 4 (via nodes A, 2, and C) in Panel C of the diagram above would be assigned a value of 3.25, 2.71, 6, or 1 if the arithmetic mean, geometric mean, maximum, or minimum was used, respectively. The global coefficient based on the four methods would be 0.6494, 0.6806, 0.6, and 0.7778, respectively. Given that all 4-paths contain a tie weight of 6 and the topology of the network is identical to the binary two-mode network in Panel A, the coefficient attained with the maximum method is equal to the binary coefficient. The increases in the coefficients, when other methods for defining 4-path values are used, are a reflection of the fact that the closed 4-paths have relatively stronger ties than the open 4-paths. The various explanations given in Opsahl and Panzarasa (2009) for the differences between the methods for defining 2-paths values are also applicable in the case of 4-path values.
Clustering coefficients for two-mode networks: Local coefficient
As noted above, the local global clustering coefficient suffers from a number of issues if applied to projections of two-mode data. The local clustering coefficient can be redefined in a similar vein as the global clustering coefficient for two-mode networks. While the one-mode local coefficient was based on 2-paths centered on a focal node, this could be extended to 4-paths centered on a focal node in two-mode networks. This would imply that the first and last nodes of the path are of the same node set as the focal node. For example, node 3 in the network shown in Panel A of the diagram above is in the center of two 4-paths, where node 1 can be seen as the first node, and nodes 2 and 4 as the last ones. A 4-path does not exist from node 2 to node 4 (via node C, node 3, and node C) as node C is part of it twice. If such a path would be included in the measure, it would be closed by definition. Formally, the local clustering coefficient for two-mode networks is:
This coefficient has similar properties as the global coefficient. First, for each node, the coefficient varies between 0 and 1 as the numerator and denominator are positive numbers, and the numerator is a subset of the denominator. Second, all 4-paths are closed in a fully connected network, and therefore, coefficient is equal to 1. Third, if ties are randomly placed in the network, the expected value of the local clustering coefficient is . By using the generalization of the local clustering coefficient for weighted one-mode networks (Barrat et al., 2004), the two-mode local clustering coefficient can be generalized for weighted two-mode networks. By using the same 4-path values as the global coefficient, it is possible to differentiate them. In turn, it is possible to define a local clustering coefficient for weighted two-mode networks that share the same properties as the binary one, and is roughly equal to the binary one if weights are randomly assigned to ties.
Want to try with your data
The global and local clustering coefficients for two-mode networks can be calculated using tnet, and the reinforcement coefficient can be calculated using the R-code below. First, you need to download and install tnet in R. Then, you need to create an edgelist of your network (see the data structures in tnet for two-mode networks). The commands below show how to load Davis’ (1940) Southern Women data and how to calculate the three coefficients.
# Load tnet library(tnet) # Load Davis' (1940) Southern Women Dataset data(tnet) net <- Davis.Southern.women.2mode net <- as.tnet(net, type="binary two-mode tnet") # Calculate the reinforcement coefficient (Robins and Alexander, 2004) reinforcement_tm(net) # Calculate the global coefficient (Opsahl, 2012) clustering_tm(net) # Calculate the local coefficient (Opsahl, 2012) clustering_local_tm(net)
Code used on this page
The code below can be used to reproduce the first diagram showing the Norwegian Interlocking directorate.
# Load tnet
library(tnet)
# Download Norwegian interlocking directorate network
net <- read.table("http://boardsandgender.com/data/net2m/net2m_2009-08-01.txt")
net <- as.tnet(cbind(net[,2], net[,1]), type="binary two-mode tnet")
# Download gender information for colouring
net.gender <- read.table("http://boardsandgender.com/data/data_people.txt", header=TRUE)
net.gender <- as.character(net.gender[,"gender"])
net.gender[net.gender=="1"] <- "#000000"
net.gender[net.gender=="2"] <- "#FFFFFF"
# Project onto a one-mode network
net.projected <- projecting_tm(net, method="binary")
# Load as igraph object
net.igraph <- tnet_igraph(net.projected)
# Set gender as a vertex attribute
net.igraph <- set.vertex.attribute(net.igraph, "color", value=net.gender[1:vcount(net.igraph)])
# Extract main component
net.components <- clusters(net.igraph)
net.main.component <- induced.subgraph(net.igraph, which(net.components$membership==which.max(net.components$csize)))
# Plot network
plot(net.main.component, layout=layout.fruchterman.reingold.grid, vertex.size=3, vertex.label=NA)
References
Barrat, A., Barthelemy, M., Pastor-Satorras, R., Vespignani, A., 2004. The architecture of complex weighted networks. Proceedings of the National Academy of Sciences 101 (11), 3747-3752.
Borgatti, S. P., Everett, M. G., 1997. Network analysis of 2-mode data. Social networks 19, 243-269.
Burt, R.S., 1992. Structural holes. Harvard University Press, Cambridge, MA.
Burt, R.S., 2005. Brokerage and closure: An introduction to social capital. Oxford University Press, Oxford, UK.
Holland, P. W., Leinhardt, S., 1970. A method for detecting structure in sociometric data. American Journal of Sociology 76, 492-513.
Latapy, M., Magnien, C., Del Vecchio, N., 2008. Basic notions for the analysis of large two-mode networks. Social Networks 30(1), 31-48.
Lind, P.G., González, M.C., Herrmann, H.J., 2005. Cycles and clustering in bipartite networks. Physical Review E 72, 056127.
Luce, R. D., Perry, A. D., 1949. A method of matrix analysis of group structure. Psychometrika 14 (1), 95-116.
Newman, M. E. J., 2001. Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E 64, 016132.
Opsahl, T., 2013. Triadic closure in two-mode networks: Redefining the global and local clustering coefficients. Social Networks 35, doi:10.1016/j.socnet.2011.07.001.
Opsahl, T., Panzarasa, P., 2009. Clustering in weighted networks. Social networks 31, 155-163.
Peng, W., Sharpe, K., Robins, G.L., Pattison, P.E., 2009. Exponential random graph (p*) models for affiliation networks. Social Networks 31, 12–25.
Robins, G., Alexander, M., 2004. Small worlds among interlocking Directors: Network structure and distance in bipartite graphs. Computational and Mathematical Organization Theory 10 (1), 69–94.
Seierstad, C., Opsahl, T., 2011. For the few, not the many? The effects of affirmative action on presence, prominence, and social capital of women directors in Norway. Scandinavian Journal of Management 27 (1), 44-54.
Snijders, T. A. B., 2001. The statistical evaluation of social network dynamics. Sociological Methodology 31, 361-395.
Watts, D. J., Strogatz, S. H., 1998. Collective dynamics of “small-world” networks. Nature 393, 440-442.
Wasserman, S., Faust, K., 1994. Social Network Analysis: Methods and Applications. Cambridge University Press, New York, NY.
Zhang, P., Wang, J., Li, X., Li, M., Di, Z., Fan, Y., 2008. Clustering coefficient and community structure of bipartite networks. Physica A 387, 6869–6875.
1. John T Scholz | February 27, 2012 at 9:02 pm
John T Scholz | February 27, 2012 at 9:02 pm
For a weighted 2-mode network, the command “clustering_local_tm(net2)” produces the follow output for the first lines:
node lc lc.am lc.gm lc.ma lc.mi
1 1 0.5789474 0.5756300 0.5568930 0.5759220 0.4851772
2 2 0.3787879 0.4117520 0.4336639 0.3799618 0.5025974
3 3 0.2736781 0.2912917 0.2849269 0.2914947 0.2684981
4 4 0.3240741 0.3355950 0.3482124 0.3313095 0.3813559
5 5 NaN NaN NaN NaN NaN
6 6 NaN NaN NaN NaN NaN
I assume that the columns with suffixes am, gm, ma, and mi are calculated with the arithmetic and geometric mean and the maximum and minimum value for the 4-paths, but how is the first lc column calculated? I could not find an explanation in the manual or online.
Thanks, John
2. Tore Opsahl | February 27, 2012 at 9:24 pm
Tore Opsahl | February 27, 2012 at 9:24 pm
Hi John,
Thanks for spotting a missing piece of the documentation. The second column called lc is simply the binary local clustering coefficient for two-mode networks.
Let me know if there is anything else – I will incorporate this in the upcoming version of tnet!
Best,
Tore
3. Yaseswini | March 3, 2013 at 1:08 pm
Yaseswini | March 3, 2013 at 1:08 pm
hi,
I am using the tnet package for bipartite graph analysis. Iam particularly interested in calculating the clustering coefficient of all the vertices of the network. But the documentation says that the clustering coefficient function “clustering_local_tm(net)” returns the values for the primary node set.How do i calculate the clustering coefficient of the second set? could you please explain
Thanks
Yaseswini
4. Tore Opsahl | March 3, 2013 at 9:14 pm
Tore Opsahl | March 3, 2013 at 9:14 pm
Hi Yaseswini,
You can swap the columns to get local clustering scores for the secondary nodes. For example, if you network is in an object called net which has two columns, you can simply type
net <- net[,2:1]to reverse the columns.Best,
Tore
5. Yaseswini | March 4, 2013 at 4:28 am
Yaseswini | March 4, 2013 at 4:28 am
Thanks for the reply.I shall try that.
Yaseswini
6. Yaseswini | March 5, 2013 at 6:50 am
Yaseswini | March 5, 2013 at 6:50 am
Hi Tore,
I am using the clustering_local_tm function for my two mode data which has 6029 vertices in the primary node set and 9313 vertices in the secondary node set with 57912 edges connecting them.When i tried using the function, i got an error message stating:
Error: cannot allocate vector of size 1.4 Gb
In addition: Warning messages:
1: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 4043Mb: see help(memory.size)
2: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 4043Mb: see help(memory.size)
3: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 4043Mb: see help(memory.size)
4: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 4043Mb: see help(memory.size)
can u please help me out with the problem.
Thanks
Yaseswini
7. Tore Opsahl | March 5, 2013 at 5:32 pm
Tore Opsahl | March 5, 2013 at 5:32 pm
Hi Yaseswini,
You are running out of memory.. The R-functions were written to be faster than memory efficient due to loops being really really slow in R. For larger networks, I have c++ functions that are very fast and very memory efficient; however, they require some knowledge to work. Email me and I will send them to you.
Best,
Tore
8. Jon | July 1, 2013 at 9:45 am
Jon | July 1, 2013 at 9:45 am
Hi Tore,
I was using the clustering_local_tm(net) for my two-mode-network with 700 vertices in the primery node set and 24 in the secondary node set. The resulting table contains a lot of values that are “Not a number” i.e.
Node lc
696 696 NaN
697 697 NaN
698 698 NaN
699 699 NaN
700 700 0.008620690
701 701 NaN
Is there any explanation for this?
Thanks!
9. Tore Opsahl | July 1, 2013 at 6:28 pm
Tore Opsahl | July 1, 2013 at 6:28 pm
Hi Jon,
The local clustering coefficient is undefined (or NaN in R) for isolates and nodes with only one connection. This is similar to the one-mode version as the denominator (in the one-mode coefficient) is n(n-1) where n is the number of connections. If n=1, the denominator is equal to 0, and it is not possible to divide a numerator with 0.
Hope this helps,
Tore
10. Jung, Sung Hoon | November 18, 2013 at 11:04 am
Jung, Sung Hoon | November 18, 2013 at 11:04 am
Hi Tore,
I am using your tnet package in R. I’d like to calculate two mode data which has about 1,700 vertices in the primary data and about 1,800 vertices in the secondary data. But when i tried using your tnet package in R, i got an error message, often. Such as below.
Do you have any solutions on these problems? pleas teach me your methods.
from jung sung hoon in S. KOREA
Error: cannot allocate vector of size 1.8 Gb
In addition: Warning messages:
1: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 8053Mb: see help(memory.size)
2: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 8053Mb: see help(memory.size)
3: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 8053Mb: see help(memory.size)
4: In `[.data.frame`(x, c(m$xi, if (all.x) m$x.alone), c(by.x, seq_len(ncx)[-by.x]), :
Reached total allocation of 8053Mb: see help(memory.size)
11. Tore Opsahl | November 19, 2013 at 2:26 pm
Tore Opsahl | November 19, 2013 at 2:26 pm
Hi Jung Sung Hoon,
This is a problem of using R. I have traded-off speed with memory usage (large objects vs loops). If you are using the clustering_tm-function, then there is a c++ version that is much faster and memory efficient. Email me if you need these files.
Best,
Tore
12. Jung, Sung Hoon | November 20, 2013 at 10:15 am
Jung, Sung Hoon | November 20, 2013 at 10:15 am
Thank you very much!!!
13. Abdul Waheed | December 24, 2014 at 9:41 am
Abdul Waheed | December 24, 2014 at 9:41 am
Dear Tore,
I have found local clustering coefficient for my network and in case of GM method I’m getting NA result in whole column. On the other hands I’m getting values for remaining methods. The r-project shows that produced by integer overflow. What could be reason for this. Further, is there any way to plot power-law behavior on log-log scale in tnet? I’ll remain thankful for your kind concern on this.
Kind regards
A.W.Mahesar
14. Tore Opsahl | December 24, 2014 at 2:33 pm
Tore Opsahl | December 24, 2014 at 2:33 pm
Hi A.W.,
If your tie weights are high, you might get integer overflow (especially on the 32-bit version of R). This is a limitation of R.
For an example of plotting degree distributions on log-log scales with regression line, see https://toreopsahl.com/2009/10/16/similarity-between-node-degree-and-node-strength/
Best,
Tore
15. Abdul Waheed | December 25, 2014 at 7:23 am
Abdul Waheed | December 25, 2014 at 7:23 am
Dear Tore,
Thank you very much for your kind concern and reply.
Kind regards,
A.W.Mahesar
16. Jinseok Kim | September 24, 2015 at 2:43 pm
Jinseok Kim | September 24, 2015 at 2:43 pm
Hello Tore,
I am currently using tnet for a large dataset without any problem.
Thank you again for your kind help.
I have a question.
One of my collaboration networks shows a tm clustering coefficient (0.34) higher than Newman’s clustering coefficient (0.33).
In your paper (Triadic closure in two-mode networks: Redefining the global and local clustering coefficients – Social Networks), you reported tm clustering coefficients a little higher than Newman’s measure for random networks of Scientific Collaboration and Norwegian Directors networks (Table 1).
So, I think it is possible that a tm clustering coefficient can be higher than Newman’s coefficient in a network.
But the problem is that I can not figure out what network structure or cases can cause this.
Could you please provide any clue or insight to me?
Best regards,
Jinseok Kim
17. Tore Opsahl | September 26, 2015 at 12:37 pm
Tore Opsahl | September 26, 2015 at 12:37 pm
Hi Jinseok,
Thank you for reaching out. I’m glad you find tnet helpful.
There are two aspects of all clustering coefficients: a numerator and a denominator. While most explanations are solely focused on the numerator, the denominator is key when comparing the ratio. It is true that one-mode projections “over-count” the number of triangles as a single secondary node with three or more nodes generate an “automatic triangle”. In the paper, I argued that these triangles do not represent triadic closure as this notion is related to an existing triplet (e.g., A->B->C) that are responsible for a heightened probability of a closing tie (i.e., A->C). These automatic triangles increase the numerator as well as the denominator as both open and closed triplets are also counted there. The over-counting happens because all these triangles are closed.
Now, how does this compare to the two-mode clustering coefficient. First, there are a different number of 4-paths in these networks (i.e., the denominator is different). This number is not necessarily smaller than the denominator in the projected one-mode network as multiple secondary nodes among primary nodes can create more 4-paths than projected triplets. For example, the ties:
A->1->B
A->2->B
B->3->C
create two 4-paths from A to C (whereas the projected one-mode network only would have a single triplet):
A->1->B->3->C
A->2->B->3->C
In essence, if it is more common among closed 4-paths to have multiple secondary nodes than the bias of automatic triangles in the projected one-mode network, the two-mode clustering coefficient could be higher than the one computed on the projected one-mode network.
Hope this helps,
Tore
18. Jinseok Kim | September 27, 2015 at 2:18 am
Jinseok Kim | September 27, 2015 at 2:18 am
Tore,
Thank you so much for your kind reply.
You answered my question.
Best regards,
Jinseok
19. Fareena | March 18, 2017 at 11:23 am
Fareena | March 18, 2017 at 11:23 am
Hi Tore
I have a bipartite network with about 2000 rows and 2 columns in a csv file ( partially shown here)
RefID,Attributes
17562,24
17573,67
17574,82
17580,55
17613,40
17613,57
17613,85
17616,24
17630,75
17632,9
17643,13
17672,25
17711,40
17733,40
17733,57
17733,85
17791,43
17797,24
17807,41
17818,13
17901,32
17936,67
17941,78
17977,82
17977,21
18001,19
18011,23
18012,34
18050,81
18057,81
18070,79
18088,83
Would I be able to obtain the unique clusters of the attributes from the second column such as {40,57,85}, {82,21} so that I can compare them and check whether they exist across other csv files using tnet. I have done one mode projections of the bipartite matrix and have obtained edge lists for the 2 columns separately but I seem to lose valuable information of clusters that have more than 2 attributes. Hence working with a two mode matrix seems more sensible. I would be grateful for any pointers in the right direction.
20. Tore Opsahl | March 18, 2017 at 10:22 pm
Tore Opsahl | March 18, 2017 at 10:22 pm
Hi Fareena,
I am not entirely sure what your goal is. You could compute the distance among attributes using the distance function with gc_only=FALSE.
Best,
Tore
21. Fareena | March 18, 2017 at 11:20 pm
Fareena | March 18, 2017 at 11:20 pm
Thank you , Tore for your response. I will try to work on this. Just to clarify -My aim is to extract attribute information from at least 30 tables similar to the one above and the analyse these values. I want to consider only those RefIDs which have 2 or more attributes and exclude those with single attr values.
When I obtain the one mode projection of the above csv file, I get an edge list with vertices V1 and V2 with weights such as v1 40 v2 57 w 2. This results in loss of data as in some cases there are 3 vertices , for example ( RefID 17613 has 3 attributes 40 57 and 85).
22. mparke26 | July 5, 2017 at 10:05 pm
mparke26 | July 5, 2017 at 10:05 pm
Quick question: are these global coefficients (reinforcement & clustering) usable for highly fragmented bipartite networks (numerous isolated components of varying sizes)? The more I think about it, the less I think they will provide meaningful measures of a highly fragmented network large real-world network. I am pretty new to network analysis, so I’d appreciate the opinion of someone who is more knowledgeable on the subject.
If these coefficients are not compatible with fragmented bipartite networks, are there any alternative measures that might be useful here?
If anyone knows, please share!
23. Tore Opsahl | July 6, 2017 at 3:41 am
Tore Opsahl | July 6, 2017 at 3:41 am
Hi Matt,
Those measures should work on fragmented networks. The one metric which is sensitive to fragmentation is closeness (or at least the standard version of it: https://toreopsahl.com/2010/03/20/closeness-centrality-in-networks-with-disconnected-components/)
Good luck!
Tore
24. mparke26 | July 6, 2017 at 6:10 pm
mparke26 | July 6, 2017 at 6:10 pm
Thanks Tore!
25. Elena Stasewitsch | July 19, 2018 at 10:48 am
Elena Stasewitsch | July 19, 2018 at 10:48 am
Hi Tore,
I just read your paper on the Small-World Phenomenon and was wondering if you provided the script for calculating the randomized networks somewhere and in particular how you created the figure for the
distributions of values (from tie reshuffled networks and weight reshuffled networks (based on 1,000 randomizations))? Did you use tnet or any other program? I would be very happy about a few hints:)
Best,
Elena
26. Tore Opsahl | July 19, 2018 at 8:30 pm
Tore Opsahl | July 19, 2018 at 8:30 pm
Hi Elena,
I did indeed use tnet (and the c++ version of the clustering functions). To get the code, check with my coauthor Antoine as he continued the work after I left academia.
Best,
Tore
27. Elena Stasewitsch | July 21, 2018 at 5:10 am
Elena Stasewitsch | July 21, 2018 at 5:10 am
Hi Tore,
thank you for your quick reply, I already contacted him and I hope he will write back soon. Thank you so much for your help and this awesome website:)
Best,
Elena
28. wp1310024334 | February 16, 2024 at 11:19 pm
wp1310024334 | February 16, 2024 at 11:19 pm
Hi Tore,
I’m Toshitaka in Japan. Thank you for your excellent research and analysis program.
I am interested in the “clustering_local_tm” in “tnet”.
In the following two-mode network, actors 1, 2, and 3 are closed by events A, B, and C. If I calculate the clustering coefficient by tnet, it is 1.
On the other hand, actors 4, 5, and 6 are closed by event D, but the clustering coefficient cannot be calculated by tnet.
Is there an option in the “clustering_local_tm” of “tnet” to assume that actors 4, 5, and 6 are also closed?
eventA eventB eventC eventD
actor1 1 1 0 0
actor2 1 0 1 0
actor3 0 1 1 0
actor4 0 0 0 1
actor5 0 0 0 1
actor6 0 0 0 1
Toshitaka
29. Tore Opsahl | February 18, 2024 at 1:24 pm
Tore Opsahl | February 18, 2024 at 1:24 pm
Hi Toshitaka,
The two mode clustering coefficient is undefined for actors 4 to 6 in your example as their two mode degree is less than 2 — ie they do not have an opportunity to be connectors. If you do not want to apply this assumption, you could always compute the degree using degree_tm-function first.
Tore