Projection
 Two-mode networks are rarely analysed in their original form. Although this is preferable, few methods exist for that purpose. As such, these networks are often transformed into one-mode networks (only one type of nodes) to be analysed. This procedure is often referred to as projection. Projection is done by selecting one of the sets of nodes and linking two nodes from that set if they were connected to the same node (of the other kind). This process is illustrated for the blue nodes of this diagram. For example, node E and node F would be connected as they have ties to a common red node. Projected networks often contains large fully-connected cliques as ties are formed among all nodes in the one-mode network that were connected to a single other node in the two-mode structure. These networks are often binary one-mode networks (admittedly, I am guilty of analysing binary projections, e.g., Seierstad and Opsahl, 2011); however, information from the two-mode network can be incorporated as tie weights. The usefulness of this approached has been illustrated by Padrón et al. (2011). By studying a number of networks, the found that this approach “allow us to make more realistic predictions on the potential competitive or facilitative interactions among species of one set (e.g. plants) that share species of the other (e.g. flower visitors or dispersers).”
Two-mode networks are rarely analysed in their original form. Although this is preferable, few methods exist for that purpose. As such, these networks are often transformed into one-mode networks (only one type of nodes) to be analysed. This procedure is often referred to as projection. Projection is done by selecting one of the sets of nodes and linking two nodes from that set if they were connected to the same node (of the other kind). This process is illustrated for the blue nodes of this diagram. For example, node E and node F would be connected as they have ties to a common red node. Projected networks often contains large fully-connected cliques as ties are formed among all nodes in the one-mode network that were connected to a single other node in the two-mode structure. These networks are often binary one-mode networks (admittedly, I am guilty of analysing binary projections, e.g., Seierstad and Opsahl, 2011); however, information from the two-mode network can be incorporated as tie weights. The usefulness of this approached has been illustrated by Padrón et al. (2011). By studying a number of networks, the found that this approach “allow us to make more realistic predictions on the potential competitive or facilitative interactions among species of one set (e.g. plants) that share species of the other (e.g. flower visitors or dispersers).”
Binary Two-mode Networks
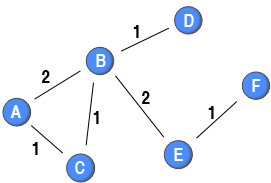 Traditionally, the ties in projected one-mode networks do not have weights attached to them. However, recent empirical studies of two-mode networks has created a weighted one-mode network by defining the weights as the number of co-occurrences (e.g., the number of events two individuals have co-attended or the number of papers that two authors had collaborated on). To formally describe this method and ease the comparison among the different methods introduced in this post, this method can be formalised as:
Traditionally, the ties in projected one-mode networks do not have weights attached to them. However, recent empirical studies of two-mode networks has created a weighted one-mode network by defining the weights as the number of co-occurrences (e.g., the number of events two individuals have co-attended or the number of papers that two authors had collaborated on). To formally describe this method and ease the comparison among the different methods introduced in this post, this method can be formalised as: where where
is the weight between node
and node
, and
is the nodes of the other kind that node
and node
are connected to, their co-occurrences (e.g., the red nodes in the above diagram). In the sample network, the tie between node A and node B has a weight of 2 as these two nodes have connections to two common red nodes, whereas the tie between node A and node C has only a weight of 1 as these nodes have connections to merely one common red node.
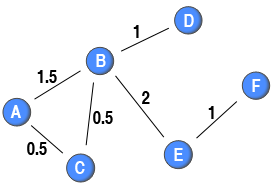 Newman (2001) extended this procedure while working with scientific collaboration networks. He argued that the social bonds among scientist collaborating with few others on a paper were stronger than the bonds among scientists collaborating with many on a paper. He proposed to discount for the size of the collaboration by defining the weights among the nodes using the following formula:
Newman (2001) extended this procedure while working with scientific collaboration networks. He argued that the social bonds among scientist collaborating with few others on a paper were stronger than the bonds among scientists collaborating with many on a paper. He proposed to discount for the size of the collaboration by defining the weights among the nodes using the following formula: where
is the number of authors on paper
(e.g., the number of blue nodes connected to the red node
). In the context of scientific collaboration networks, this implies that if two scientists who only write a single paper together with no other co-authors get a weight of 1 (e.g., node B and node D). Moreover, if two scientists have written two papers together without any co-author, the weight of their tie would be 2 (e.g., node B and E). However, if the two scientists have a co-author, the weight on the tie between them is 0.5 (e.g., node A and node C). A more complicated example is the tie between node A and node B in the diagram. They have written two papers together: one without any other co-author and one with node C as a co-author. The first paper would give their tie a weight of 1, and the second tie would add 0.5 to the weight of this tie. Therefore, the weight attached to their tie is 1.5. By discounting for the number of blue nodes attached to the same red node, this methods creates a one-mode projection in which the strength of a node is equal to the number of ties originating from that node in the two-mode network (e.g. the sum of weights attached to ties originating from node A in the one-mode projection is 2, and node A is connected to two red nodes).
Weighted Two-mode Networks
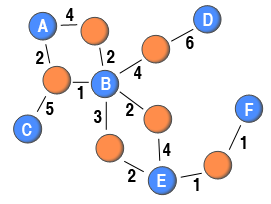 So far, the methods has only dealt with binary two-mode networks. However, weighted two-mode networks also exist. For example, in online forums, where the node sets are users and threads (or topics), and a tie between two nodes is established if a user posts a message to a thread, it is possible to differentiate the two-mode ties based on the number of messages or characters posted by users to a specific thread. Another example is purchasing behaviour where the node set are people and goods, and the weight can be defined in terms of number of purchases. Similarily to weighted one-mode networks, the data used for analysis is richer if the tie weights are included in the two-mode network than if they are discarded. A weighted two-mode network is illustrated in the diagram to the right.
So far, the methods has only dealt with binary two-mode networks. However, weighted two-mode networks also exist. For example, in online forums, where the node sets are users and threads (or topics), and a tie between two nodes is established if a user posts a message to a thread, it is possible to differentiate the two-mode ties based on the number of messages or characters posted by users to a specific thread. Another example is purchasing behaviour where the node set are people and goods, and the weight can be defined in terms of number of purchases. Similarily to weighted one-mode networks, the data used for analysis is richer if the tie weights are included in the two-mode network than if they are discarded. A weighted two-mode network is illustrated in the diagram to the right.
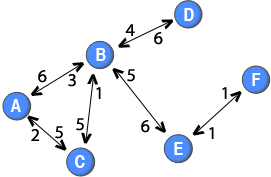
Note: The weights of the directed ties are placed close to the node that the ties originate from. For example, the tie from node B to node D has a weight of 4, whereas the tie from node D to node B has a weight of 6.
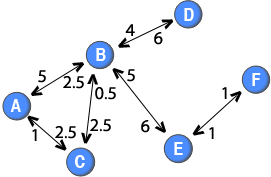 In a similar spirit as the method used by Newman (2001), it is also possible to discount for the number of nodes when projecting weighted two-mode networks. For example, it could be argued that if many online users post to a thread, their ties should be weaker than if there were few people posting to the thread. A straight forward generalisation is the following function:
In a similar spirit as the method used by Newman (2001), it is also possible to discount for the number of nodes when projecting weighted two-mode networks. For example, it could be argued that if many online users post to a thread, their ties should be weaker than if there were few people posting to the thread. A straight forward generalisation is the following function: . This formula would create a directed one-mode network in which the out-strength of a node is equal to the sum of the weights attached to the ties in the two-mode network that originated from that node. For example, node C has a tie with a weight of 5 in the two-mode network and an out-strength of 5 in the one-mode projection.
Want to test it with your data?
Binary two-mode networks
The following code requires a binary two-mode network to be listed in an edgelist format with two columns named i and p. The i column refers to the nodes you would like to keep in the one-mode projection (e.g., the blue nodes), and the p column refers to the nodes you would like to discard (e.g., the red nodes). The binary two-mode network in the diagram above can be loaded using the following function.
net <- cbind( i=c(1,1,2,2,2,2,2,3,4,5,5,5,6), p=c(1,2,1,2,3,4,5,2,3,4,5,6,6))
The one-projections highlighted above can be created using the following code:
# Load tnet library(tnet) # Binary one-mode projection projecting_tm(net, method="binary") # Simply the number of common red nodes projecting_tm(net, method="sum") # Newman's method projecting_tm(net, method="Newman")
Weighted two-mode networks
The following code requires a weighted two-mode network to be listed in an edgelist format with three columns named i, p, and w. The i column refers to the nodes you would like to keep in the one-mode projection (e.g., the blue nodes), the p column refers to the nodes you would like to discard (e.g., the red nodes), and the w column must be the weight of the ties. The weighted two-mode network in the diagram above can be loaded using the following function.
net.w <- cbind( i=c(1,1,2,2,2,2,2,3,4,5,5,5,6), p=c(1,2,1,2,3,4,5,2,3,4,5,6,6), w=c(4,2,2,1,4,3,2,5,6,2,4,1,1))
The one-projections highlighted above can be created using the following code:
# Load tnet library(tnet) # Simply the sum of weights towards common red nodes projecting_tm(net.w, method="sum") # Generalisation of Newman's method projecting_tm(net.w, method="Newman")
References
Davis, A., Gardner, B. B., Gardner, M. R., 1941. Deep South. University of Chicago Press, Chicago, IL.
Newman, M. E. J., 2001. Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E 64, 016132.
Opsahl, T., 2013. Triadic closure in two-mode networks: Redefining the global and local clustering coefficients. Social Networks 35, doi:10.1016/j.socnet.2011.07.001.
Padrón, B., Nogales, M., Traveset, A., 2011. Alternative approaches of transforming bimodal into unimodal mutualistic networks. The usefulness of preserving weighted information. Basic and Applied Ecology, doi:10.1016/j.baae.2011.09.004.
Seierstad, C., Opsahl, T., 2011. For the few, not the many? The effects of affirmative action on presence, prominence, and social capital of women directors in Norway. Scandinavian Journal of Management 27 (1), 44-54.
1. Snehal | September 13, 2012 at 11:16 pm
Snehal | September 13, 2012 at 11:16 pm
Hi Tore
Great resource particularly for weighted 2-mode networks.
I am working with 3-mode networks and figured that I’ll have to convert (project) it to 2 modes to get some measures. Can you suggest any literature that deals with this conversion? Right now I am just replicating the 2->1 mode procedures. Your comments/suggestions are greatly appreciated.
Thanks!
2. Tore Opsahl | September 17, 2012 at 6:33 pm
Tore Opsahl | September 17, 2012 at 6:33 pm
Hi Snehal,
Thanks! I have not directly worked with 3-mode networks, so I haven’t given too much thought. I think this is an exciting area of new research with many unanswered questions. Do you project from 3-mode to two-mode by connecting primary and secondary nodes connected to the same tertiary nodes? What do you do with existing connections between primary and secondary nodes?
Best,
Tore
3. Snehal | September 17, 2012 at 10:35 pm
Snehal | September 17, 2012 at 10:35 pm
Thanks Tore for your reply. Yes, at this point, I am considering that primary and secondary nodes are connected if they share a tertiary node. And, the number of common tertiary nodes is captured in the tie strength. Interestingly, given the nature of my original data, primary and secondary nodes are only connected to a tertiary node (women attending event, women having some sort of expertise). So I guess the 2nd question hasn’t been an issue with this dataset.
Best
Snehal
4. Mary | October 31, 2012 at 5:48 am
Mary | October 31, 2012 at 5:48 am
Hi Tore,
Thank you for your article. I was looking for something which would be illustrative and precise. And your article is just that. I am currently working on a two mode bipartite network. Do you know how I can use neighborhood measures like Common Neighbors in the bipartite setting? Thanks
5. Tore Opsahl | October 31, 2012 at 1:49 pm
Tore Opsahl | October 31, 2012 at 1:49 pm
Hi Mary,
Glad you found it useful. I am not entirely sure what you are referring to when you mention neighborhood measures. However, if you would like to calculate the number of common nodes between primary nodes in a two-mode network, you can use the projecting_tm(net, method=”sum”). This function will produce pairs of primary nodes (columns 1 and 2) with a third column with the number of common nodes.
Best,
Tore
6. David Fisher | January 17, 2013 at 2:24 pm
David Fisher | January 17, 2013 at 2:24 pm
Hi Tore,
Great blog, this section in particular is my go-to guide to two-mode networks.
I am interesting in using the geometric mean, instead of the sum, to calculate the strength of an interaction between 2 nodes when projecting a 2-mode network onto a one-mode network.
The strength of the interaction between nodes A and B in the above example (leaving out Newman’s correction for now) would be 2 in this case (or 4.24 if you sum the weights directed at nodes they share before taking the geometric mean).
Do you think this is sensible, and can it be done in tnet?
Cheers
David
7. Tore Opsahl | January 21, 2013 at 8:37 pm
Tore Opsahl | January 21, 2013 at 8:37 pm
Hi David,
Great suggestion. It does sound like a good idea as differences in tie weights would discount the tie strength. I haven’t implemented this type of projection method for two-mode networks; however, if you connect with me by email, this code can easily be created.
Best,
Tore
8. Martin Grandjean | March 12, 2013 at 9:46 pm
Martin Grandjean | March 12, 2013 at 9:46 pm
What a great description! I’ve just published a “state of my research” on my blog (www.martingrandjean.ch/analyse-de-reseau-cartographier-lactivite-de-la-societe-des-nations/) and as a historian (passionate about network analysis but without maths studies) I was wondering how to justify theoretically this “projection”. Thank you for these keys!
9. Tore Opsahl | March 12, 2013 at 10:43 pm
Tore Opsahl | March 12, 2013 at 10:43 pm
Hi Martin,
Great to see more open science being done! I’m not sure there is a specific theory I can point you too regarding projects as there is so many. For example, two-mode networks with people and events can be projected due to assumed temporal geographical co-location. However, I believe it really depends on your context which theories of connectedness would apply.
Best,
Tore
10. Abdul Waheed | November 14, 2014 at 3:18 pm
Abdul Waheed | November 14, 2014 at 3:18 pm
Dear Tore,
Hi,
I’m working on two-mode networks. In above post the tnet command for extended weighted network formula is not given.
Only binar,sum and newman is given. Kindly if possible can you please tell me the command for last method.
I’ll remain thankful.
Kind regard,
A.W.Mahesar
11. Tore Opsahl | November 15, 2014 at 12:23 am
Tore Opsahl | November 15, 2014 at 12:23 am
The default method is “sum”; in the text, the binary method is not shown as all weights would be one, and the two diagrams below each two-mode network is based on the “sum” and “Newman” methods, respectively.
Hope this helps,
Tore
12. Abdul Waheed | November 15, 2014 at 9:07 am
Abdul Waheed | November 15, 2014 at 9:07 am
Thanks dear for your kind concern. Further, for projection based on the Newman extended formula for weighted 2-mode network which command can be use.
Regards,
13. Tore Opsahl | November 15, 2014 at 3:18 pm
Tore Opsahl | November 15, 2014 at 3:18 pm
Abdul,
You are simply using the projection_tm-function with method=”Newman” and the network being a weighted two-mode network: see line #8 in the fourth and final code block above.
Tore
14. Abdul Waheed | November 15, 2014 at 4:10 pm
Abdul Waheed | November 15, 2014 at 4:10 pm
Dear Opsahl,
Thanks for your kind information. Now I got the answer.
Kind regards,
15. Andy | March 23, 2015 at 9:23 pm
Andy | March 23, 2015 at 9:23 pm
Hi Tore,
Congrats for your website, I find it very helpful and thanks for sharing your knowledge.
I have two questions, and I would be grateful if you could answer : Can I calculate centrality measures on the projected network?
Also, is it possible, in a multi-mode network for the one set of actors to be directed(and if yes, can I use your package to turn it into a projected network)?
Thanks,
Andy
16. Tore Opsahl | March 23, 2015 at 11:34 pm
Tore Opsahl | March 23, 2015 at 11:34 pm
Hi Andy,
I’m glad you’re finding the site useful.
1) To calculated node centrality on a projected two-mode network, you must use the one-mode metrics. See https://toreopsahl.com/tnet/weighted-networks/node-centrality/
2) Two-mode networks (tnet is limited to two-mode networks for multi-mode networks) are generally undirected, or directional in one way only. To the best of my knowledge, there are no general directional two-mode metrics.
Good luck,
Tore
17. Sandra | January 4, 2016 at 10:11 pm
Sandra | January 4, 2016 at 10:11 pm
Hi Tore,
I am doing a one-mode projection of a weighted biparite network. I noticed that in the projection, the weights for 1-2 are different than from 2-1 and so on (see below), even though my network is not directional.
I am using the command: projecting_tm(mynet, method=”sum”). I tried adding directed=NULL but I get an “unused argument” error. Is there a way to specify that the network is undirected?
i j w
1 1 2 37
2 1 3 1
3 1 4 52
4 1 5 62
5 2 1 38
6 2 3 1
7 2 4 50
8 2 5 43
9 3 1 1
10 3 2 1
11 3 4 2
12 3 5 1
13 4 1 202
14 4 2 157
15 4 3 2
16 4 5 299
17 5 1 185
18 5 2 110
19 5 3 1
20 5 4 289
Thanks in advance!!!
18. Tore Opsahl | January 5, 2016 at 12:52 am
Tore Opsahl | January 5, 2016 at 12:52 am
Hi Sandra,
The method creates a two ties — one in each direction — between primary nodes that a secondary node; however, the tie weights are not guaranteed to be symmetric. In the weighted two-mode network example above, node A has a total tie weight of 6 towards nodes shared with node B; however, node B only have a total tie weight of 3 with common nodes.
Hope this helps,
Tore
19. Sandra | January 5, 2016 at 2:24 pm
Sandra | January 5, 2016 at 2:24 pm
Hi Tore,
Thank you so much for your fast response! The data above was the one-dimension projection. Below is the original weighted two-mode bipartite network where i represents frugivore species, j fruit species, and w frequencies of interaction (number of individuals of species i consuming fruit j).
In the one-dimension projection of frugivores I would expect symmetrical weights because frugivores are indirectly connected by the fruit they share irrespective of direction. For example in the projection (using SUM method) I got: species1-species2 = 37, species2- species1 = 38. I’m I misinterpreting something? Thanks again!
Original bipartite network:
*Edges
i j w
1 7 3
1 9 5
1 15 7
1 16 1
1 17 21
1 18 1
1 19 2
1 20 2
1 21 4
1 24 1
1 25 3
1 28 3
1 30 1
1 37 12
1 38 1
2 6 8
2 9 1
2 10 4
2 15 3
2 17 26
2 18 4
2 20 3
2 29 1
2 33 4
2 38 1
2 41 2
3 34 1
3 38 1
4 6 13
4 7 21
4 8 14
4 9 15
4 10 24
4 11 2
4 12 2
4 13 8
4 14 14
4 15 41
4 16 28
4 17 33
4 18 11
4 19 1
4 20 19
4 21 22
4 22 2
4 23 2
4 24 17
4 25 11
4 26 1
4 28 1
4 30 2
4 31 1
4 32 3
4 34 1
4 35 1
4 36 2
4 38 1
4 39 2
4 40 1
4 42 3
4 43 1
4 44 4
5 7 32
5 8 19
5 9 11
5 10 13
5 12 1
5 13 9
5 14 3
5 15 21
5 16 1
5 17 5
5 18 14
5 20 43
5 21 18
5 22 5
5 23 14
5 24 33
5 25 7
5 26 1
5 27 2
5 28 6
5 29 2
5 30 23
5 31 1
5 35 3
5 36 2
5 37 2
5 38 1
5 40 1
5 43 1
5 44 1
One dimension projection (SUM method from t-net):
1 2 37
1 3 1
1 4 55
1 5 65
2 1 38
2 3 1
2 4 50
2 5 43
3 1 1
3 2 1
3 4 2
3 5 1
4 1 223
4 2 157
4 3 2
4 5 299
5 1 217
5 2 110
5 3 1
5 4 289
20. Tore Opsahl | January 6, 2016 at 5:45 am
Tore Opsahl | January 6, 2016 at 5:45 am
Hi Sandra,
I was referring to the sample graphs in the figures above. Specifically, the weighted two-mode network (fourth figure on this page) and the corresponding weighted one-mode network using the sum-method (fifth figure on this page).
For your dataset, primary nodes 1 and 2 share the following secondary nodes: 9, 15, 17, 18, 20, and 38. The sum of node 1’s ties with these common nodes is 37 (5+7+21+1+2+1), and the sum of node 2’s tie with the common nodes is 38 (1+3+26+4+3+1). As such, the projection-function using the sum-method creates two ties between nodes 1 and 2:
i j w
1 2 37
2 1 38
Hope this further clarifies what the function does.
Best,
Tore
21. Sandra | January 6, 2016 at 3:15 pm
Sandra | January 6, 2016 at 3:15 pm
Hello Tore,
Thank you very much for the explanation! This makes sense!
Sandra
22. VIVEK | August 3, 2016 at 12:24 am
VIVEK | August 3, 2016 at 12:24 am
Hi Tore, WONDERFUL POST. I was wondering, if I want to relabel the entities, for example in my case the column i is an “Person” and column j is a “post” I was wondering how can I get back to the original labels back as cbind is converting them to numeric labels.
Thanks for your help
23. Tore Opsahl | August 3, 2016 at 2:14 am
Tore Opsahl | August 3, 2016 at 2:14 am
Hi Vivek,
If you are asking about the column names, you can set these by using the colnames-functions. For example:
Good luck,
Tore
24. officeinc | August 3, 2016 at 2:47 am
officeinc | August 3, 2016 at 2:47 am
Hi Tore thanks for quick reply,
what I am asking is as follows. I have my weighted network as below,
Names Posts frequency
A Politics 2
A travel 40
B Politics 5
B Money 2
C Politics 24
C Money 23
D Politics 20
When I use the cbind method, it creates them into numeric columns
1 1 2
1 2 40
2 1 5
2 3 2
3 1 2
3 3 24
4 1 23
and so on…
I was wondering if after projecting I can get back the matrix as
A B 54
B C 36
D A 68 etc.
and also if i get the centrality measures with the lables (A, B, C) rather than 1, 2, 3
Hope this provides better clarity.
Thanks a lot
Vivek
25. Tore Opsahl | August 3, 2016 at 3:50 am
Tore Opsahl | August 3, 2016 at 3:50 am
Hi Vivek,
It seems that your data is a data.frame with the first two columns being factors. When a factor is transformed into a matrix, the labels are lost. To get the labels (or levels as they are called), you can write the following code is your network is an object called net:
Good luck!
Tore
26. Marion | February 20, 2017 at 8:10 am
Marion | February 20, 2017 at 8:10 am
Hi Tore,
In the case of the Binary Two-mode Networks, why do the links (B,C) and (A,C) are valued 0.5 instead of 0.33 each ?
I would assume that in the case of the paper written by author A, B and C;
each links will be valued 0.33 so that the sum of (B,C), (A,C) and (A,B) for this paper will be one.
Thanks for your help !
Marion
27. Tore Opsahl | February 22, 2017 at 1:27 am
Tore Opsahl | February 22, 2017 at 1:27 am
Hi Marion,
The algorithm works so that the node strength is equal to the number of papers written. See Mark Newman’s article for more details on his method.
Best,
Tore
28. vivek | February 22, 2017 at 9:38 am
vivek | February 22, 2017 at 9:38 am
Hi Tore, Thanks for replying to my earlier doubt. One of the question I have is regarding, 2-mode networks weighted degree centrality. I was wondering what is the best way to normalize the value obtained? Will sum of the total weighted degree centralities be a good option for that. To provide an example, if we have two nodes (after converting to 1-mode networks) and w1 and w2 are the weighted degree centralities respectively. Should I use w1/(w1+w2) as the normalised degree centrality ? is there a better way to find the normalized measure? Thanks for your help.
29. Tore Opsahl | February 25, 2017 at 12:59 am
Tore Opsahl | February 25, 2017 at 12:59 am
Hi Vivek,
The normalization of tie weights is highly dataset dependent. Nonetheless, certain metrics are defined to normalize weights for a person (e.g., Burt’s, 1992, constraint metric defines p_ij as w_ij / w_i++). I have chosen to stay away from such assumptions, and leave it up the research to determine the appropriate handling of tie weights for their dataset.
Best,
Tore
30. Bert | August 17, 2017 at 10:52 am
Bert | August 17, 2017 at 10:52 am
Hi Tore,
1. Our two-mode network is weighted and some of the weights are negative. Do you see any problems projecting such a network into a weighted one-mode network?
2. Let A be an m x n matrix describing a two-mode network. Do I understand it correctly that–somewhat simplifying–the tnet command projecting_tm calculates A’A (and/or AA’) and then sets the diagonal entries to zero?
31. Tore Opsahl | August 29, 2017 at 2:55 am
Tore Opsahl | August 29, 2017 at 2:55 am
Hi Bert,
I haven’t worked with negative weights, so I am not entire sure of the code’s behavior in these instances. Projection works by multiplying a matrix with its transpose; however, tnet does it through merges due to using an internal edgelist representation of networks. The tie weights is based on the selected method.
Best,
Tore