Clustering in Weighted Networks
 A fundamental measure that has long received attention in both theoretical and empirical research is the clustering coefficient. This measure assesses the degree to which nodes tend to cluster together. Evidence suggests that in most real-world networks, and in particular social networks, nodes tend to create tightly knit groups characterised by a relatively high density of ties (Feld, 1981; Friedkin, 1984; Heider, 1946; Holland and Leinhardt, 1970; Louch, 2000; Snijders, 2001; Snijders et al., 2006; Watts and Strogatz, 1998). In real-world networks, this likelihood tends to be greater than the average probability of a tie randomly established between two nodes (Holland and Leinhardt, 1971; Wasserman and Faust, 1994; Watts and Strogatz, 1998).
A fundamental measure that has long received attention in both theoretical and empirical research is the clustering coefficient. This measure assesses the degree to which nodes tend to cluster together. Evidence suggests that in most real-world networks, and in particular social networks, nodes tend to create tightly knit groups characterised by a relatively high density of ties (Feld, 1981; Friedkin, 1984; Heider, 1946; Holland and Leinhardt, 1970; Louch, 2000; Snijders, 2001; Snijders et al., 2006; Watts and Strogatz, 1998). In real-world networks, this likelihood tends to be greater than the average probability of a tie randomly established between two nodes (Holland and Leinhardt, 1971; Wasserman and Faust, 1994; Watts and Strogatz, 1998).
Traditionally, the two versions of the clustering coefficient developed for testing the tendency of nodes to cluster together into tightly knit groups are the global clustering coefficient (Doreian, 1969) and the local clustering coefficient (Watts and Strogatz, 1998). The global version was designed to give an overall indication of the clustering in the network, whereas the local gives an indication of the embeddedness of single nodes.
The Global Clustering Coefficient
The global clustering coefficient is based on triplets of nodes. A triplet is three nodes that are connected by either two (open triplet) or three (closed triplet) undirected ties. A triangle consists of three closed triplets, one centred on each of the nodes. The global clustering coefficient is the number of closed triplets (or 3 x triangles) over the total number of triplets (both open and closed). The first attempt to measure it was made by Luce and Perry (1949). This measure gives an indication of the clustering in the whole network (global), and can be applied to both undirected and directed networks (often called transitivity, see Wasserman and Faust, 1994, page 243). However, it cannot be applied to weighted networks.
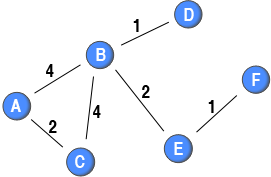 For this sample network, the binary global clustering coefficient would be 3 over 9, or 0.33. The closed triplets are B→A←C; A→B←C; A→C←B; whereas the open triplets are A→B←D; A→B←E; C→B←D; C→B←E; D→B←E; B→E←F. As can be seen from the sample network, the strongest ties are inside the triangle. This is not captured by the binary coefficient as the weights are not considered.
For this sample network, the binary global clustering coefficient would be 3 over 9, or 0.33. The closed triplets are B→A←C; A→B←C; A→C←B; whereas the open triplets are A→B←D; A→B←E; C→B←D; C→B←E; D→B←E; B→E←F. As can be seen from the sample network, the strongest ties are inside the triangle. This is not captured by the binary coefficient as the weights are not considered.
A generalisation of the global clustering coefficient to weighted networks was proposed by Opsahl and Panzarasa (2009). This generalisation required a triplet value to be defined, and then, divided the value of closed triplets on the value of all triplets. The generalised clustering coefficient produces the same result as the binary global clustering coefficient when it is applied to a binary network. This is because all triplets have the same value, irrespective of the method used to calculate triplet values. In addition, the generalised coefficient has the same properties as C. Moreover, if weights are randomly assigned in the network, the weighted clustering coefficient is equal to the binary coefficient.
Four methods was suggested for defining a triplet value: the arithmetic mean, geometric mean, maximum, and minimum of the two tie weights that make up the triplet. The arithmetic mean is simply the average of the tie weights. This method is insensitive to differences between the two tie weights as an extreme value can have a major impact on the triplet value. The geometric mean of the weights attached to the two ties overcomes some of the sensitivity issues as a triplet made up by a tie with a low value and a tie with a high value will have a lower value than if the arithmetic mean were used. In addition, it is possible to use two extreme methods to deal with differences in tie weights. The maximum method takes the highest value of the two weights, and will make a triplet with a strong tie and a weak tie equal to a triplet with two strong ties. Conversely, the minimum method takes the lowest value of the two weights, and make triplets with a strong tie and a weak tie equal to triplets with two weak ties. The table below highlights the differences between the methods of defining the triplet value (adopted from Chapter 2 of my thesis).

It is vital to use an appropriate method for defining the value of a triplet as this impacts on the outcome of the coefficient. The method should be chosen based on the research question as well as the way in which the strength of the ties are operationalised into weights. For example, in a network where the weights correspond to the level of flow, and a weak tie would act as a bottleneck, the minimum method might be most appropriate to use.
 To highlight a potential use of the binary and generalised global clustering coefficient, it is possible to see whether stronger triplets are more likely to be closed than weaker ones. This can be done by comparing the weighted version (
To highlight a potential use of the binary and generalised global clustering coefficient, it is possible to see whether stronger triplets are more likely to be closed than weaker ones. This can be done by comparing the weighted version () to the binary version calculated on a binary network where all ties with a weight greater than 0 is set to present (
). In so doing, the two coefficients are calculated on the same network structure. The only difference is that the former takes the weights of ties into consideration. If the ratio between the weighted clustering coefficient and the binary coefficient is higher than 1, it can be argued that triplets made up by strong ties are more likely to be closed than triplets made up by weak ties (
). On the contrary, it can be argued that triplets made up by weak ties are more likely to be closed than those made up by strong ties if the ratio is less than 1.
To empirically investigate whether strong ties are more likely to be part of closed triplets than weak ties, I have used the tnet networks. For simplicity, the arithmetic mean method is used for defining triplet values.
| Network | Nodes | Ties | ||||
|---|---|---|---|---|---|---|
| 1 | Freeman’s EIES network (time 1) | 48 | 695 | 0.7627 | 0.7702 | 1.0099 |
| 2 | Freeman’s EIES network (time 2) | 48 | 830 | 0.8131 | 0.8214 | 1.0102 |
| 3 | Freeman’s EIES network (messages) | 32 | 460 | 0.6386 | 0.7378 | 1.1555 |
| 4 | Consulting (advice) | 46 | 879 | 0.6932 | 0.7130 | 1.0285 |
| 5 | Consulting (value) | 46 | 858 | 0.6764 | 0.6852 | 1.0131 |
| 6 | Research team (advice) | 77 | 2228 | 0.6848 | 0.7127 | 1.0408 |
| 7 | Research team (awareness) | 77 | 2326 | 0.6785 | 0.6957 | 1.0253 |
| 8 | C.elegans’ neural network | 306 | 2345 | 0.1818 | 0.2364 | 1.3009 |
| 9 | US airport network | 500 | 2980 | 0.3514 | 0.4765 | 1.3562 |
| 10 | Newman’s scientific collaboration network | 16730 | 47594 | 0.3596 | 0.3178 | 0.8838 |
As it is possible to see from the above table, 9 of the 10 networks have a weighted clustering coefficient that is higher than the traditional clustering coefficient. This suggest that triangles generally are, in fact as suggested by Simmel (1923[1950]), made up by strong ties. The tenth network, which shows the opposite result, is different from the other nine networks. It is a one-mode projection of a two-mode network, whereas the first nine networks are native one-mode networks. This feature could explain the observed result as the one-mode projection is constructed in a way that penalises the weight of ties within triangles, or more specifically, large fully connected cliques (Newman, 2001). See the projecting two-mode networks.
Local Clustering Coefficient
The local clustering coefficient is based on ego network density or local density (Scott, 2000; Uzzi and Spiro, 2005; Watts and Strogatz, 1998). For a node, this is the fraction of the number of present ties over the total number of possible ties between the node’s neighbours. Therefore, the outcome ranges between 0 and 1: 0 if no ties exist between the neighbours, and 1 if all possible ties exists.
 For the sample network, nodes A and C would get a score of 1 since all possible ties among their neighbours are present, whereas node E would get a score of 0 since none of the possible ties among its neighbours are present. Nodes D and F would not get a value since the number of possible ties among their neighbours is 0 (if a node has less than 2 neighbours, the coefficient is undefined). For node B, one out of six possible ties is present, so the coefficient is 1/6 or 0.1667.
For the sample network, nodes A and C would get a score of 1 since all possible ties among their neighbours are present, whereas node E would get a score of 0 since none of the possible ties among its neighbours are present. Nodes D and F would not get a value since the number of possible ties among their neighbours is 0 (if a node has less than 2 neighbours, the coefficient is undefined). For node B, one out of six possible ties is present, so the coefficient is 1/6 or 0.1667.
The main advantage of this version of the clustering coefficient is that a score is assigned to each node (local measure). This enables researchers to study associations between the coefficient and other nodal properties (e.g. Panzarasa et al., 2009) and perform regression analyses with the observations being the nodes of a network (e.g. Uzzi and Lancaster, 2004). However, this version of the clustering coefficient suffers from three major limitations (see Opsahl and Panzarasa, 2009, for a longer discussion). First, its outcome does not take into consideration the weight of the ties in the network. Second, the local clustering coefficient cannot be calculated on directed networks. Third, a negative correlation with degree is often found in real-world networks. This is due to the fact that it is “easier” for a node with two neighbours to get a score of 1 (only one tie is need) than for a node with 10 neighbours (45 ties must be present).
Recently, there have been a number of attempts to extend the local clustering coefficient to the case of weighted networks (Barrat et al., 2004; Lopez-Fernandez et al., 2004; Onnela et al., 2005; Zhang and Horvath, 2005). I will focus on Barrat et al. (2004) measure. They proposed a generalisation of the local clustering coefficient to weighted networks by taking the weights explicitly into account. First, they assigned a triplet value to each triplet in the network based on the arithmetic mean (normal average). Then for each node, they summed the value of the closed triplets that were centred on the node and divided it by the total value of all triplets centred on the node. Similarily to the weighted global clustering coefficient, triplet values do not have to be the arithmetic mean, but could also be the geometric mean, maximum, and minimum of the two tie weights.
For the above sample network, all the nodes, except node B, would get the same outcome as the binary analysis. Nodes A and C get a value of 1 because all their neighbours are connected, whereas node E get a value of 0 because none of its neighbours are connected. Nodes D and F do not have a value as they have less than 2 neighbours. Node B does not get the same value as it has more than 2 neighbours, and is the centre of both open and closed triplets. The weighted local clustering coefficient is between 0.18 and 0.36, while the binary one is 0.17. The increased weighed coefficient is a reflection of node B’s strong ties being directed towards neighbours that are themselves connected. The table below highlights the local clustering coefficients.
| Node | Weighted local clustering coefficient using | ||||
|---|---|---|---|---|---|
| arithmetic mean | geometric mean | maximum method | minimum method | binary version | |
| 1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 2 | 0.24 | 0.27 | 0.18 | 0.36 | 0.17 |
| 3 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 4 | NaN | NaN | NaN | NaN | NaN |
| 5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 6 | NaN | NaN | NaN | NaN | NaN |
Want to test it with your data?
The clustering coefficients can be calculated using tnet. First, you need to download and install tnet in R. Then, you need to create an edgelist of your network (see the data structures in tnet for weighted one-mode networks). The commands below show how the edgelist for the sample network here can manually be entered, and how to calculate the clustering coefficients.
# Load tnet
library(tnet)
# Manually enter the sample network
net <- cbind(
i=c(1,1,2,2,2,2,3,3,4,5,5,6),
j=c(2,3,1,3,4,5,1,2,2,2,6,5),
w=c(4,2,4,4,1,2,2,4,1,2,1,1))
# Compute the global clustering coefficients
clustering_w(net, measure=c("bi", "am", "gm", "ma", "mi"))
# Compute the local clustering coefficients
clustering_local_w(net, measure=c("bi", "am","gm","ma","mi"))
The code used to create the above table was:
# Load tnet
library(tnet)
# Load networks
data(tnet)
net <- list()
net[[1]] <- Freemans.EIES.net.1.n48
net[[2]] <- Freemans.EIES.net.2.n48
net[[3]] <- Freemans.EIES.net.3.n32
net[[4]] <- Cross.Parker.Consulting.net.info
net[[5]] <- Cross.Parker.Consulting.net.value
net[[6]] <- Cross.Parker.Manufacturing.net.info
net[[7]] <- Cross.Parker.Manufacturing.net.aware
net[[8]] <- celegans.n306.net
net[[9]] <- USairport.n500.net
net[[10]] <- Newman.Condmat.95.99.net.1mode.wNewman
# Calculate values
output <- data.frame(CGT0=NaN, Cw=NaN, Ratio=NaN)
for(i in 1:length(net))
output[i,c("CGT0","Cw")] <- clustering_w(net[[i]], measure=c("bi","am"))
output[,"Ratio"] <- output[,"Cw"]/output[,"CGT0"]
References
Barrat, A., Barthelemy, M., Pastor-Satorras, R., Vespignani, A., 2004. The architecture of complex weighted networks. Proceedings of the National Academy of Sciences 101 (11), 3747-3752. arXiv:cond-mat/0311416
Burt, R. S., 1992. Structural Holes: The Social Structure of Competition. Harvard University Press, Cambridge, MA.
Coleman, J. S., 1988. Social capital in the creation of human capital. American Journal of Sociology 94, S95-S120.
Colizza, V., Pastor-Satorras, R., Vespignani, A., 2007. Reaction-diffusion processes and metapopulation models in heterogeneous networks. Nature Physics 3, 276-282. arXiv:cond-mat/0703129
Doreian, P., 1969. A note on the detection of cliques in valued graphs. Sociometry 32 (2), 237-242.
Feld, S. L., 1981. The focused organization of social ties. American Journal of Sociology 86, 1015-1035.
Friedkin, N. E., 1984. Structural cohesion and equivalence explanations of social homogeneiety. Sociological Methods and Research 12, 235-261.
Granovetter, M., 1973. The strength of weak ties. American Journal of Sociology 78, 1360-1380.
Heider, F., 1946. Attitudes and cognitive organization. Journal of Psychology 21, 107-112.
Holland, P. W., Leinhardt, S., 1970. A method for detecting structure in sociometric data. American Journal of Sociology 76, 492-513.
Holland, P. W., Leinhardt, S., 1971. Transitivity in structural models of small groups. Comparative Group Studies 2, 107-124.
Louch, H., 2000. Personal network integration: Transitivity and homophily in strong-tie relations. Social Networks 22, 45-64.
Luce, R. D., Perry, A. D., 1949. A method of matrix analysis of group structure. Psychometrika 14 (1), 95-116.
Opsahl, T., Panzarasa, P., 2009. Clustering in weighted networks. Social Networks 31 (2), 155-163.
Panzarasa, P., Opsahl, T., Carley, K. M., 2009. Patterns and dynamics of users’ behavior and interaction: Network analysis of an online community. Journal of the American Society for Information Science and Technology 60 (5), 911-932.
Scott, J., 2000. Social Network Analysis: A Handbook. Sage Publications, London, UK.
Simmel, G., 1950. The Sociology of Georg Simmel (KH Wolff, trans.). Free Press, New York, NY.
Snijders, T. A. B., 2001. The statistical evaluation of social network dynamics. Sociological Methodology 31, 361-395.
Snijders, T. A. B., Pattison, P. E., Robins, G. L., Handcock, M. S., 2006. New specifications for exponential random graph models. Sociological Methodology 35, 99-153.
Uzzi, B., Lancaster, R., 2004. Embeddedness and price formation in the corporate law market. American Sociological Review 69, 319-344.
Uzzi, B., Spiro, J., 2005. Collaboration and creativity: The small world problem. American Journal of Sociology 111, 447-504.
Wasserman, S., Faust, K., 1994. Social Network Analysis. Cambridge University Press, Cambridge, MA.
Watts, D. J., Strogatz, S. H., 1998. Collective dynamics of “small-world” networks. Nature 393, 440-442.
1. Jinie Pak | July 17, 2013 at 9:19 pm
Jinie Pak | July 17, 2013 at 9:19 pm
Hi Tore,
I have a question about tnet clustering coefficient function.
My data is weighted one-mode network (edge list).
I tried to get clustering coefficient score for this network, but I kept getting the following error messages:
Error in clustering_local_w(data) : Network is not undirected!
Measure is not defined from directed networks.
In addition: Warning message:
In as.tnet(net, type = “weighted one-mode tnet”) :
There were self-loops in the edgelist, these were removed
Can you point out what I am doing wrong?
Thank you!
2. Tore Opsahl | July 19, 2013 at 12:49 am
Tore Opsahl | July 19, 2013 at 12:49 am
Jinie,
This happens because you have ties from a node to the same node (i.e., where the i column is equal to the j column).
Tore
3. Barbara | January 29, 2015 at 3:20 pm
Barbara | January 29, 2015 at 3:20 pm
Hi,
Is there an option to disable this behavior? I.e. to preserve and count self-loops in your “alpha” measure?
Thanks
4. Tore Opsahl | January 31, 2015 at 11:39 pm
Tore Opsahl | January 31, 2015 at 11:39 pm
Hi Barbara,
There isn’t one, but there is a trick for the degree-function (for closeness and betweenness, self-loops do not matter): add a sink node as the degree function is local. See the code below.
Good luck,
Tore
# Load tnet library(tnet) # Generate random net net <- rg_w(n=5, arcs=0.2) # Add self-loops with a weight equal to node id net <- rbind( net, data.frame(i=1:5, j=1:5, w=1:5)) net <- net[order(net[,1], net[,2]),] # So the code for you would be: # Find the largest node id and add 1 sinkNode <- max(c(net[,"i"],net[,"j"]))+1 # Copy network netSinkNode <- net # Redirect receivers of self-loops to new node (replace "j" with "i" if in-degree) netSinkNode[netSinkNode[,"i"]==netSinkNode[,"j"],"j"] <- sinkNode # Compute degree out <- degree_w(netSinkNode, measure=c("degree","output","alpha"), alpha=0.5) # Remove net node out <- out[out[,"node"]!=sinkNode,]5. Theo | October 24, 2013 at 9:57 am
Theo | October 24, 2013 at 9:57 am
Hi Tore,
A question about the global clustering function.
I apply the function to small weighted networks. I normally keep the network data in a matrix format. I have converted the matrixes to edgelists before I applied the global clustering coefficients function but I noticed that the results are the same as when I apply the function directly to the data in the matrix format. Is there any special reason why I should keep converting the data?
Best regards!
Theo
6. Tore Opsahl | October 24, 2013 at 2:14 pm
Tore Opsahl | October 24, 2013 at 2:14 pm
Hi Theo,
Thank you for your comment!
All tnet functions can be applied to edgelists and matrices. Specifically, the as.tnet-function is also run first by the functions if it hasn’t manually be run already. This function makes a number of assumptions based on the properties of the supplied data object:
As good practice, I would always recommend running as.tnet and specifying the type-parameter manually after loading your network. For example:
net.matrix <- read.table(…)
net <- as.tnet(net.matrix, type="weighted one-mode tnet")
clustering_w(net)
Best,
Tore
7. Theo | October 25, 2013 at 10:19 am
Theo | October 25, 2013 at 10:19 am
Hi Tore,
Thank you for your quick reply and a very useful answer! Since I work with square matrices tnet transforms them to weighted one-mode tnet. I will check the type-parameter to be sure.
Since I work with small networks I want to keep track of different nodes. The fact that nodes are given a number when the matrix is converted to an edgelist makes this more difficult. Is there an easy way to get node ID’s from a matrix to the edgelist?
Best regards,
Theo
8. Tore Opsahl | October 26, 2013 at 2:04 pm
Tore Opsahl | October 26, 2013 at 2:04 pm
Hi Theo,
I am not entire sure what you mean by node ids as this information is not contained in the matrix of ties except for row/column numbers. The as.tnet function takes the row/column number as the id by using which applied to a matrix with arr.ind equal to TRUE (line 40 in the as.tnet-function: net 0, arr.ind = TRUE), net[net > 0]) ).
If you would like consistent id numbers, I suggest explicitly create edgelists with the numbers.
Best,
Tore
9. Eduarda | March 24, 2014 at 4:40 pm
Eduarda | March 24, 2014 at 4:40 pm
Hi Tore,
a have a question about the output of clustering_w. The result is supposed to be in a range between 0 and 1 right? I’m applying this to a non directed network represented by a symmetric matrix of 44 vertex and weighted edges. All the values are non negative.
Using the comand:
clu2<-clustering_w(O2,measure=c("am", "gm", "ma", "mi"))
I'm getting
clu2
am gm ma mi
1.986471 4.322812 7.850534 1.288835
Do you have any idea of what am I doing wrong?
10. Tore Opsahl | March 25, 2014 at 1:51 am
Tore Opsahl | March 25, 2014 at 1:51 am
Hi Eduarda,
The clustering coefficient should indeed be between 0 and 1. Could you send me your data and code? Then I will have a look at it.
Best,
Tore
11. Lorenzo | July 26, 2014 at 6:12 pm
Lorenzo | July 26, 2014 at 6:12 pm
Hello. I’d like to leave you a question.
On en.wikipedia, at a certain point (https://en.wikipedia.org/wiki/Clustering_coefficient#Global_clustering_coefficient), it says that Watts and Strogatz defined the GCC as (as I have understood) the average of the LCCs. But I’ve done some tries and the method of the triplets and the method of the average of the LCCs are not equivalent.
How is it possible?
12. Tore Opsahl | July 26, 2014 at 6:33 pm
Tore Opsahl | July 26, 2014 at 6:33 pm
Hi Lorenzo,
There are multiple ways of defining the overall level of clustering in a network. The global clustering coefficient which is defined as 3x triangles over triples has been around for a long time (all the way back to Luce and Perry, 1949). In their 1998 paper, Watts and Strogatz introduced the local clustering coefficient, which is a node level metric (studying the number of ties among neighbors were not a new idea, see Granovetter, 1973, Coleman, 1988, and Burt, 1992). To get an overall coefficient, Watts and Strogatz took a simple average of the node-level scores. By using a simple average, you get a different answer than the global clustering coefficient if the nodes do not all have the same number of ties. To get the same answer, you can use a weighted average where each node is weighted by the number of triplets they are the center of, n*(n-1) (see code below).
Hope this helps and clarifies!
Tore
13. Lorenzo | July 27, 2014 at 11:29 am
Lorenzo | July 27, 2014 at 11:29 am
Thank you very much for the immediate response!
14. Rose | August 28, 2014 at 7:16 pm
Rose | August 28, 2014 at 7:16 pm
Hi Tore,
I have a geographic distance matrix (distance of one household to another). I have taken the inverse distance, so households located closer to each other have stronger weights. All the households have the same number of connections. I am interested in calculating network level measures, instead of individual nodes. I have 100 20×20 (dimension) matrices. I have tried calculating the global clustering coefficient for each matrix after I have transformed each matrix to an edge list. However, the global clustering coefficient I get for all the matrices are 1, which is not what I would expect because of the different weights for each matrix.
Also, would you be able to recommend other network level measures? I know you have discussed some of the issues of calculating somehow aggregate measures, but I was just wondering maybe it may not be a big issue if all households are connected to the same number of households anyway and that the network structure is fixed (household location)? Thanks in advance.
Sincerely,
Rose
15. Tore Opsahl | August 30, 2014 at 12:02 am
Tore Opsahl | August 30, 2014 at 12:02 am
Hi Rose,
The clustering metrics all become 1 when the network is fully connected as would be in your case. Have you thought about the weighted rich-club?
Let me know how you end up analyzing your network!
Tore
16. Rose | August 30, 2014 at 12:38 am
Rose | August 30, 2014 at 12:38 am
Hi Tore,
Ohh, that was what I was guessing. Thanks for confirming.
About the weighted rich-club suggestion, I did see the R commands you posted. I also read through you paper and I think it is a good measure for what I am looking for. I followed what you have posted (which may be a wrong application for me), but I think I may be doing something wrong and hence not getting the weighted rich club coefficient. Here is the command I used:
out <- weighted_richclub_w(edge_b2, rich="k", reshuffle="links", NR=1000, seed=1)
where edge_b2 is an edgelist for one of my neighborhoods. Is that how I do it? Cause I'm not getting the answer. Sorry if this seems like an easy question but I couldn't figure it out as of now. Thanks so much!
Sincerely,
Krisha
17. Tore Opsahl | August 30, 2014 at 12:39 am
Tore Opsahl | August 30, 2014 at 12:39 am
Krisha,
In the case of a fully connected network, I would use the weight reshuffling. Let me know how it works!
Tore
18. Sérgio Tomás | September 18, 2014 at 2:24 pm
Sérgio Tomás | September 18, 2014 at 2:24 pm
hi,
I’m a masters student in Lisbon University, faculty of human kinetics, and the masters is in High performance training. I’m using clustering coefficients and graph densities. i would like you to send me the pdf documents for the compilation of my thesis.
Regards,
19. Tore Opsahl | September 19, 2014 at 12:38 pm
Tore Opsahl | September 19, 2014 at 12:38 pm
Hi Sergio,
Send me an email and I will forward you the PDFs.
Tore
20. siri | January 12, 2015 at 6:59 am
siri | January 12, 2015 at 6:59 am
hi sir..after finding the cluster coefficient..how to divide the network based on the clustering coefficient..
21. Tore Opsahl | January 13, 2015 at 2:16 am
Tore Opsahl | January 13, 2015 at 2:16 am
Hi Siri,
The clustering coefficient is not a community structure algorithm.
Good luck,
Tore
22. Narisong Huhe | January 21, 2015 at 6:33 am
Narisong Huhe | January 21, 2015 at 6:33 am
Hi Tore,
Thank you so much for making tnet available. I’m an assistant professor of political science, and working on a project on the relationship between international trade and democracy. Since bilateral trade has different impacts on the two parties involved (depending on their size of economy), my data is directed and weighted. Is there any way to calculate local clustering coefficient in this case?
best,
Naris
23. Tore Opsahl | January 24, 2015 at 5:40 pm
Tore Opsahl | January 24, 2015 at 5:40 pm
Hi Naris,
The local clustering coefficient is only defined for undirected network. I would look into the structural holes theory by Ron Burt. Specifically, the constraint metric might be the appropriate one for you.
Best,
Tore
24. Akhlaq Ahmad | August 4, 2016 at 7:49 am
Akhlaq Ahmad | August 4, 2016 at 7:49 am
Hi Tore,
I was confused about the local clustering coefficient of node B that is 1/6. There is one tie between neighbours A and C so the numerator is 1, How we get 6 at the denominator?
25. Tore Opsahl | August 4, 2016 at 12:14 pm
Tore Opsahl | August 4, 2016 at 12:14 pm
Hi Akhlaq,
The denominator in the binary local clustering coefficient is the number of triplets centered on the focal node (B). These triples are A-B-C, A-B-D, A-B-E, C-B-D, C-B-E, and D-B-E. It can also be calculated as N*(N-1)/2 when N the number of neighbors in undirected networks.
Hope this helps,
Tore
26. Akhlaq Ahmad | August 4, 2016 at 3:17 pm
Akhlaq Ahmad | August 4, 2016 at 3:17 pm
Thanks a lot. I got it.
Date: Thu, 4 Aug 2016 12:14:52 +0000
To: akhlaq1973_pk@hotmail.com
27. narisong | January 29, 2015 at 10:20 pm
narisong | January 29, 2015 at 10:20 pm
Hi Tore,
Thank you so much for the comments. I was also trying to use Burt’s c as a proxy of local clustering of trade. Yet, there are still two issues. First, in Burt’s conceptualization, the network seems undirected as well. So it might be problematic to use it in the case of bilateral trade. Second, the trade network generally is a complete network. So there is few, if any, structural holes in the network.
Recently, I’ve read an article on the subject. How do you think about their approach? Thank you so much!
McAssey, Michael P., and Fetsje Bijma. forthcoming. “A Clustering Coefficient for Complete Weighted Networks.” Network Science
28. Tore Opsahl | January 31, 2015 at 11:42 pm
Tore Opsahl | January 31, 2015 at 11:42 pm
Hi Naris,
Burt’s constraint metric is created for directed networks (see Burt, 1992, chapter 2); however, this metric or any local clustering coefficient becomes irrelevant if you have a complete network. You may want to set a cut-off and remove ties below this.
Best,
Tore
29. Juan Ricardo M | May 27, 2015 at 2:09 pm
Juan Ricardo M | May 27, 2015 at 2:09 pm
Hi Tore¡. Thanks for your website. It´s been useful for my because I´m doing a work about city networks with this tools. I have a question about the maximum and minimun method measure of the global cluster coefficient. I think that, in t-net in R, the maximim correspond to minimun method and viceversa, because I compute the data of the example presented in this page manually and I found that, with the maximun method, the global cluster is 0.375, while with the minimun method the global cluster is 0.5. Regards.. Juan
30. Tore Opsahl | May 30, 2015 at 3:14 pm
Tore Opsahl | May 30, 2015 at 3:14 pm
Hi Juan,
Glad you have found the site useful!
The maximum and minimum methods for the global clustering coefficient do not refer to the maximum or minimum value of the coefficient. They are methods for calculating the triplet value. See the table with the two black-and-white triplets. It is important to recognize that the triplet method affect both the numerator and the denominator of the coefficient. The minimum method produces lower numerator and denominator than the maximum one (except if all tie weights are equal; binary network).
You might also think about the different values in terms of the value assigned to heterogeneous triplets (i.e., the two ties weights are different). The greater the difference between the tie weights, the greater the impact of changing the method. I would recommend that you hack the code for the clustering coefficient to return the numerator and denominator separately. The code below will print these values.
Best,
Tore
# Load tnet library(tnet) # Get network and name it net for simplicity data(tnet) net <- celegans.n306.net # Compute two-mode clustering when the women are primary nodes clustering_w(net, measure=c("bi","am","gm","ma","mi")) # Updated function clustering_w_ND <- function (net, measure = "am") { if (is.null(attributes(net)$tnet)) net <- as.tnet(net, type = "weighted one-mode tnet") if (attributes(net)$tnet != "weighted one-mode tnet") stop("Network not loaded properly") if (length(measure) < 1) stop("You must specify a measure") for (i in 1:length(measure)) if (!(measure[i] %in% c("bi", "am", "gm", "ma", "mi"))) stop("You must specify a correct measure") net <- net[order(net[, "i"], net[, "j"]), ] dimnames(net)[[2]] <- c("i", "j", "wij") tmp <- net dimnames(tmp)[[2]] <- c("j", "k", "wjk") tmp <- merge(net, tmp) index <- tmp[, "i"] != tmp[, "k"] tmp <- tmp[index, ] tmp <- tmp[, c("i", "k", "wij", "wjk")] dimnames(net)[[2]] <- c("i", "k", "wik") net[, "wik"] <- TRUE tmp <- merge(tmp, net, all.x = TRUE) tmp[is.na(tmp[, "wik"]), "wik"] <- FALSE index <- as.logical(tmp[, "wik"]) tmp <- tmp[, c("wij", "wjk")] output <- data.frame( numerator = rep(0, length(measure)), denominator = 0, coefficient = 0) for (i in 1:length(measure)) { if (measure[i] == "am") { tvalues <- rowSums(tmp) } else if (measure[i] == "gm") { tvalues <- sqrt(tmp[, "wij"] * tmp[, "wjk"]) } else if (measure[i] == "ma") { tvalues <- pmin.int(tmp[, "wij"], tmp[, "wjk"]) } else if (measure[i] == "mi") { tvalues <- pmax.int(tmp[, "wij"], tmp[, "wjk"]) } else if (measure[i] == "bi") { tvalues <- rep(1, nrow(tmp)) } else { stop("measure incorrectly specified") } output[i,1] <- sum(tvalues[index]) output[i,2] <- sum(tvalues) output[i,3] <- output[i,1]/output[i,2] } rownames(output) <- measure return(output) } # Get results of new function clustering_w_ND(net, measure=c("bi","am","gm","ma","mi"))31. Hang Xiong | June 17, 2015 at 10:53 am
Hang Xiong | June 17, 2015 at 10:53 am
Hi Tore,
Your website is very useful to learn the characteristics of weighted networks. Thanks a lot for your effort.
I do not quite understand how the weighted global clustering coefficient is calculated, specifically, how the weights are taken into account. In the original paper, you calculate the C_ω,gm of two sampled networks (Figure 1 (a) and (b)), the results are 0.44 and 0.23 respectively. Can you please explain how the results are calculated?
I have a network (denoted as NET) consisting of a number of fully connected clusters, i.e., all nodes are connected to one another in the clusters. The weights are normalized, so the values of weights are between 0 and 1. I obtain clustering_w(NET) = 1. I’m wondering how to interpret the result. This is obvious if NET was a binary network. Since a weight will be add when calculating the weighted clustering coefficient, I though the result should not 1 any more. Can you please guide me here?
Many thanks,
Hang
32. Tore Opsahl | June 17, 2015 at 11:57 am
Tore Opsahl | June 17, 2015 at 11:57 am
Hi Hang,
Glad you find the site useful.
The clustering coefficient will always be 1 for fully connected networks. This is because all triplets are closed, and thus the triplet value (however calculated) of all triplets are part of the numerator. As the denominator is the sum of all triplet values, the numerator is the same as the denominator, hence a ratio of 1.
Hope this helps,
Tore
33. Hang Xiong | June 18, 2015 at 8:51 am
Hang Xiong | June 18, 2015 at 8:51 am
Hi Tore,
Thank you for your reply.
I tied to calculate the clustering coefficients of the two sampled networks (Figure 1 (a) and (b)), but get slightly different results. I am wondering if I do not understand the algorithm correctly. The following are my calculation.
Figure 1 (a) Network where stronger triplets are closed
Closed triplets: B→A←C; A→B←C; A→C←B
value of closed triplets: 3+4+3=10
Open triplets: A→B←D; A→B←E; C→B←D; C→B←E; D→B←E; B→E←F
value of open triplets: 2.5+3+2.5+3+1.5+1.5=14
Clustering coefficient: 10/(10+14)=0.4167
Figure 1 (b) Network where weaker triplets are closed
Closed triplets: B→A←C; A→B←C; A→C←B
value of closed triplets: 1.5+1+1.5=10
Open triplets: A→B←D; A→B←E; C→B←D; C→B←E; D→B←E; B→E←F
value of open triplets: 2.5+1.5+2.5+1.5+3+3=14
Clustering coefficient: 4/(4+14)=0.2222
Best,
Hang
34. Tore Opsahl | June 21, 2015 at 2:12 pm
Tore Opsahl | June 21, 2015 at 2:12 pm
Hi Hang,
It seems your calculations is based on arithmetic mean and the numbers reported in the paper is geometric mean.
From an undirected perspective, there are 9 triplets in these network. For Figure 1a, the triplets and their arithmetic and geometric mean-based values are:
The fractions are 10/24 = 0.4167 and 9.66/22.14 = 0.4361.
Hope this helps,
Tore
35. Abdul Waheed Mahesar | January 18, 2016 at 4:10 am
Abdul Waheed Mahesar | January 18, 2016 at 4:10 am
Hi, dear tore,
I have two questions in above given reply to Hang. First how the value of numerator in geometric mean is 9.66?
second, can you give one example for minimum and maximum method as you have given for AM and GM. I’ll always remain thankful. Regards,
36. Tore Opsahl | January 19, 2016 at 4:50 am
Tore Opsahl | January 19, 2016 at 4:50 am
Hi Abdul,
There are three closed triplets in the above example among nodes A, B, and C. These triplets have the following values on their ties {4,2}, {2,4}, and {4,4}. The geometric mean method for computing triplet values takes the square root of the product of the tie weights (e.g., sqrt(4*2)=2.83). As such, the closed triplets in the above example have the following triplet values using the GM method: 2.83, 2.83, and 4. The sum of these values is 9.66, which is the numerator.
The minimum method simply takes the minimum tie weight. In the above example, this would be: 2, 2, 4, 1, 1, 2, 2, 1, and 1. The sum of the closed ones (the first three) is 8, and the sum of all of them is 16. Hence, the clustering coefficient is 8/16 or 0.5 when using the minimum method. The maximum method is identical, but takes the maximum of the two tie weights instead.
Hope this makes it clearer,
Tore
37. Dennis Ryu | February 19, 2017 at 10:48 pm
Dennis Ryu | February 19, 2017 at 10:48 pm
Hello Tore,
I am currently conducting research on a database with undirected and weighted networks, and would greatly appreciate it if I can use your generalized clustering coefficient. As mentioned in the publication, could I possibly receive a copy of the Matlab code for doing so?
Also, is there no way to calculate the clustering coefficient for a complete graph besides setting a threshold for the edge weights and dichotomizing the network?
Thanks so much,
Dennis
38. Tore Opsahl | February 22, 2017 at 1:56 am
Tore Opsahl | February 22, 2017 at 1:56 am
Hi Dennis,
The clustering coefficient is available in R and C++. Please drop me an email for the C++ code or find the R code in tnet (which is on CRAN).
The clustering coefficient is always 1 on completed graph due to all triplets being closed.
Best,
Tore
39. Léanne B. Soucy | August 31, 2017 at 5:32 pm
Léanne B. Soucy | August 31, 2017 at 5:32 pm
Hi Tore,
This post is very useful. Thanks very much! I was wondering if there was a possibility to calculate a global clustering coefficient based on a weighted network with your package tnet, and considering the sum of the weighted edges (of triplets) instead of the arithmetic mean, geometric mean, maximum and minimum value.
Thanks very much !
Léanne
40. Tore Opsahl | August 31, 2017 at 8:10 pm
Tore Opsahl | August 31, 2017 at 8:10 pm
Hi Léanne,
Glad you’re finding it useful. If you calculate the clustering coefficient with the arithmetic mean and with the sum of triple weights, you will get the same outcome due to it being a ratio. The difference is that both the numberator and denominator will be double in the latter case.
Best,
Tore
41. Léanne B. Soucy | August 31, 2017 at 8:28 pm
Léanne B. Soucy | August 31, 2017 at 8:28 pm
Hi Tore, Thanks for the answer :)
May I please ask another question ?
I have a weighted edgelist :
Person1 Person2 Weight
[1,] “A” “B” “1”
[2,] “B” “D” “2”
[3,] “B” “C” “3”
[4,] “C” “D” “4”
Your package seems to indicate we must use a adjacency matrix.
I modified my edgelist to a adjacency matrix (name: adj_test_tr)
A B D C
A 0 1 0 0
B 1 0 2 3
D 0 2 0 4
C 0 3 4 0
as soon as I entered the command : clustering_w(adj_test_tr) i got this problem clustering_w(adj_test_tr)
Error in as.tnet(net, type = “weighted one-mode tnet”) :
Type of network not recognised
Then, I wrote to check if the format is ok
as.tnet(adj_test_tr)
Error in as.tnet(adj_test_tr) :
A node id’s below 1 is detected. The lowest possible node id is 1.
In addition: Warning message:
In as.tnet(adj_test_tr) :
Data assumed to be longitudinal tnet (if this is not correct, specify type)
My network is a simple weighted network (one-mode).
What is the proper format to ensure the clustering_w command works?
Thanks again!
Léanne
42. Tore Opsahl | September 12, 2017 at 1:45 am
Tore Opsahl | September 12, 2017 at 1:45 am
Hi Léanne,
Network data is handled as an edgelist in tnet. If you supply an adjacency matrix, it is converted. However, the node ids must be integers instead of character strings. Have a look at this page: https://toreopsahl.com/tnet/software/one-mode-data-structure/
Good luck!
Tore
43. Stefano | March 20, 2019 at 7:03 am
Stefano | March 20, 2019 at 7:03 am
Dear Tore. I have a question about how an edge’s weight is used for the local clustering coefficient. It is a cost or a capacity? I would like to compare local clustering coefficient with igraph’s local and nodal efficiencies (Latora & Melchiorri). In igraph efficiency analysis weights are seen as cost to pass through a link.
Thank you very muchs
44. Tore Opsahl | March 20, 2019 at 7:15 am
Tore Opsahl | March 20, 2019 at 7:15 am
Hi Stefano,
All links in tnet are considered strengths.
Hope this helps,
Tore
45. Lee Zhengyu | August 13, 2019 at 10:24 am
Lee Zhengyu | August 13, 2019 at 10:24 am
Dear Tore,
I have a question about the usage of as.tnet
My data is a matrix with four columes, and I want to conform it to a network which is a weighted one-mode network with tnet standards.
Then I wrote this ( b is a matrix ):
a<-as.tnet(b,type="weighted one-mode tnet")
however, I kept getting the following error message:
Error in as.tnet(b, type = "weighted one-mode tnet") :
Type of network not recognised
And if I edited it to this:
a<-as.tnet(b,type=NULL)
I got the following error message:
Error in as.tnet(b, type = NULL) : Not all node id's are integers
In addition: Warning message:
In as.tnet(b, type = NULL) :
Data assumed to be longitudinal tnet (if this is not correct, specify type)
Can you point out what I am doing wrong?
Thank you very much!
46. Tore Opsahl | August 20, 2019 at 9:11 pm
Tore Opsahl | August 20, 2019 at 9:11 pm
Hi,
The native format of tnet is edgelists. The as.tnet-function can convert matrices to edgelists, but require more than 4 nodes not to confuse with the other network types.
Best,
Tore