Shortest Paths in Weighted Networks
 Shortest paths or distances among nodes has long been a key element of network research. While the shortest paths often are not of interest in themselves, they are the key component of a number of measures. First, a popular question has been what is the average distance among people? The question became famous following Milgram’s six-degrees of separation experiment in 1967 that found that people in the US were on average 6-steps from any other person (see the introduction of my thesis for a more complete story and criticism). Second, the shortest distances have been used as an underlying metric in a number of measures, such as centrality ones (Freeman, 1978; Opsahl et al., 2010). One of these measures is betweenness, which is roughly equal to the number of shortest paths between others that pass through a node. This page is aimed at showing a generalisation of shortest path calculations for weighted networks that has the potential of being more accurate.
Shortest paths or distances among nodes has long been a key element of network research. While the shortest paths often are not of interest in themselves, they are the key component of a number of measures. First, a popular question has been what is the average distance among people? The question became famous following Milgram’s six-degrees of separation experiment in 1967 that found that people in the US were on average 6-steps from any other person (see the introduction of my thesis for a more complete story and criticism). Second, the shortest distances have been used as an underlying metric in a number of measures, such as centrality ones (Freeman, 1978; Opsahl et al., 2010). One of these measures is betweenness, which is roughly equal to the number of shortest paths between others that pass through a node. This page is aimed at showing a generalisation of shortest path calculations for weighted networks that has the potential of being more accurate.
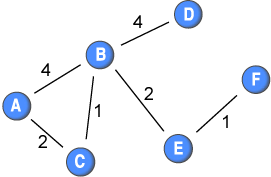
The shortest distance among nodes in a network is quite easy to calculate if you only have present or absent ties: you simply count the ties along the shortest path. If two nodes are directly connected: distance=1; and if they are not directly connected, but are connected through intermediaries, then it is the lowest number of intermediary nodes +1. For example, if the ties in this network did not have weights, the distance between node A and F would be 3. Below is a matrix showing the shortest distances among the nodes in the sample network.
A B C D E F A NA 1 1 2 2 3 B 1 NA 1 1 1 2 C 1 1 NA 2 2 3 D 2 1 2 NA 2 3 E 2 1 2 2 NA 1 F 3 2 3 3 1 NA
The average shortest distance in this network is 1.8 (the mean of the matrix, diagonal excluded).
Difficulty occurs when ties are differentiated, as they are in a weighted network. For example, the tie between node A and B has twice the strength of the tie between node A and node C. This could mean that node A has more frequent contact with node B than with node C. In turn, this could imply that node A might give node B a piece of information (or a disease) twice as likely as node C. If we are looking at the diffusion of information or diseases in a network, then the speed that it travels, and routes that it takes, are clearly affected by the weights.
Let us look at the shortest path from node C to node B. The direct connection carries a weight of 1 only; however, the indirect connection through node A is composed of stronger ties. Therefore, a piece of information (or a disease) might be transmitted quicker through node A than directly.
Dijkstra (1959) proposed an algorithm that sum the cost of connections and find the path of least resistance. For example, GPS devices uses this algorithm by assigning a time-cost to each leg of the road, and then find the route that cost least in terms of time. This algorithm can also be used in social network analysis. Newman (2001) applied it to a collaboration network of scientists by inversing the tie weights (dividing 1 by the weight). This implies that a stronger tie gets a lower cost than a weaker tie. Below is the network from above with the inverted weights:

By applying Dijkstra’s algorithm, we find that the direct connection between node C and node B has a cost of 1, whereas the indirect connection via node A has a cost of 0.75 (1/2 + 1/4). Therefore, according to this algorithm, the information will travel faster through the indirect connection. Below is a matrix showing the cost of the least costly routes among the nodes in the sample network.
A B C D E F
A NA 0.25 0.50 0.50 0.75 1.75
B 0.25 NA 0.75 0.25 0.50 1.50
C 0.50 0.75 NA 1.00 1.25 2.25
D 0.50 0.25 1.00 NA 0.75 1.75
E 0.75 0.50 1.25 0.75 NA 1.00
F 1.75 1.50 2.25 1.75 1.00 NA
The average of this matrix is 0.9833.
This application of Dijkstra’s algorithm is fine to use if we wish to identify the shortest paths; however, what does an average distance of 0.98 mean?
To this end, I suggest to “normalise” the weights by the average weight in the network. The weights would then be (before being inverted):
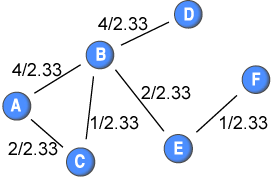
By calculating Dijkstra’s algorithm following this normalisation, we get the following shortest distances among nodes:
A B C D E F
A NA 0.5825 1.1650 1.1650 1.7475 4.0775
B 0.5825 NA 1.7475 0.5825 1.1650 3.4950
C 1.1650 1.7475 NA 2.3300 2.9125 5.2425
D 1.1650 0.5825 2.3300 NA 1.7475 4.0775
E 1.7475 1.1650 2.9125 1.7475 NA 2.3300
F 4.0775 3.4950 5.2425 4.0775 2.3300 NA
The average of this matrix is 2.29.
A unit of distance then refers to one step with the average weight in the network. The average of the matrix suggests that on average nodes are 2.29 steps with average tie weight away from each other. This measure would be comparable across networks with different ranges of tie weights.
If anyone has any suggestions for improvements, let me know!
Want to test it with your data?
This distance matrix can be calculated using tnet. First, you need to download and install tnet in R. Then, you need to create an edgelist of your network (see the data structures in tnet for weighted one-mode networks). The commands below show how the edgelist for the sample network here can manually be entered, and how to calculate the distance matrix.
# Load tnet library(tnet) # Load network net <- cbind( i=c(1,1,2,2,2,2,3,3,4,5,5,6), j=c(2,3,1,3,4,5,1,2,2,2,6,5), w=c(4,2,4,1,4,2,2,1,4,2,1,1)) # Create a binary version of the network bnet <- dichotomise_w(net) # Calculate the average distance in the binary network mean(distance_w(bnet),na.rm=T) # Calculate the average distance in the weighted network mean(distance_w(net),na.rm=T)
References
Dijkstra, E. W., 1959. A note on two problems in connexion with graphs. Numerische Mathematik 1, 269-271.
Freeman, L. C., 1978. Centrality in social networks: Conceptual clarification. Social Networks 1, 215-239.
Milgram, S., 1967. The small world problem. Psychology Today 2, 60-67.
Newman, M. E. J., 2001. Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E 64, 016132.
Opsahl, T., Agneessens, F., Skvoretz, J., 2010. Node centrality in weighted networks: Generalizing degree and shortest paths. Social Networks 32 (3), 245-251.
1. Bhupendrasinh Thakre | June 18, 2012 at 4:47 pm
Bhupendrasinh Thakre | June 18, 2012 at 4:47 pm
Awesome articles. They are helping me a lot to build my knowledge in SNA.
Thank you very much Dr. Opsahl.
2. Romana | November 1, 2013 at 2:46 pm
Romana | November 1, 2013 at 2:46 pm
First of all, thank you very much for providing all these helpful SNA resources on your blog!
I have a question concerning the tnet package.
While the command “closeness_w” returns the “raw” closeness score as well as the normalized score, the command “betweenness_w” only gives one kind of score (not normalized, I guess).
Is there a specific reason why normalization is only available for closeness centrality?
Thank you very much and best regards
Romana
3. Tore Opsahl | November 4, 2013 at 5:12 pm
Tore Opsahl | November 4, 2013 at 5:12 pm
Hi Romana,
Thank you for your comment. I am not a big fan of normalized indices as the only purpose of them is to compare nodes’ scores across networks where the number of nodes and ties vary, and to do so, it makes a couple of assumptions (e.g., scaling with n*(n-1)). This might not be appropriate. If you would like to scale the network metrics, you can simply apply the method you would like.
Best,
Tore
4. romanariegler | November 4, 2013 at 5:24 pm
romanariegler | November 4, 2013 at 5:24 pm
Thank you for your reply! So I guess if I’m just looking at one single network (no comparisons) I can just go with the non-normalized scores of betweenness and closeness.
BTW I’m sorry for my redunant comments. The first one wasn’t displayed to me until just now, so I thought something had gone wrong and posted a second time. Please feel free to delete one of them.
5. Ali | February 16, 2014 at 4:42 pm
Ali | February 16, 2014 at 4:42 pm
very good article.
Do you know any software/method can calculate the closeness centrality for a weighted directed network based on the Dijkstra’s algorithm? the commands you presented here is for unweighted network.
Thanks,
6. Tore Opsahl | February 19, 2014 at 1:12 pm
Tore Opsahl | February 19, 2014 at 1:12 pm
Hi Ali,
The tnet R package can calculate this metric for you. See the code example.
Best,
Tore
7. Walter | January 6, 2016 at 6:47 am
Walter | January 6, 2016 at 6:47 am
First, let me thank you for this very helpful blog; I found many of the entries to be highly informative and useful.
I am using iGraph for my calculations and I find the weights issue to be a bit confusing, especially after reading this entry and trying to calculate average shortest path length for networks. I assumed that in igraph the weight of an edge is a measurement of tie strength. Therefore, higher values indicate stringer connection. However, is seems that when using Dijkstra’s algorithm in Igraph, the package treats the weights of the edges as distances, looking for paths that minimize the sum of all paths between two nodes. This seems to indicate that the algorithm sees lower weights as stronger ties (ie shorter distances). My solution will be, just for this calculation to convert every edge weight x in my edge weights list to 1/x (as with the Newman reference you provided).
Did the problem I observed is right and do you think that solution a good approach?
Additionally, is this a problem only in this case (and centrality measures) but not an issue in the calculation of transitivity and modularityin igraph (for example, when using random walks).
Best,
Walter
8. Tore Opsahl | January 10, 2016 at 9:13 pm
Tore Opsahl | January 10, 2016 at 9:13 pm
Hi Walter!
Glad that you find this site useful!
Indeed the assumption that tie weights represent costs instead of strength is often leading to some incorrect findings. The distance_w-function in tnet takes the inverse of the tie weights and then utilize the igraph implementation of Dijkstra’s algorithm. I also scale the weights with the mean tie weight so that one can more easily interpret the values. I am not sure how igraph implements transitivity and modularity, so you might reach out to them directly. All the tnet functions work on the assumption of high values equal stronger connections.
Good luck!
Tore
9. Drahomir | February 28, 2016 at 8:44 pm
Drahomir | February 28, 2016 at 8:44 pm
Hi,
First thanks for a lot of useful information provided here. I had 2 questions regarding the average shortest path in weighted graph, particluary if there’s a similar way to compute Diameter of graph and also to display a distribution of shortest paths (in a way like “what is the probability of choosing a path with a certain distance if picking randomly?”)
With regards,
Drahomir
10. Tore Opsahl | March 3, 2016 at 2:46 am
Tore Opsahl | March 3, 2016 at 2:46 am
Hi Drahomir,
The diameter of a graph is simply the longest path in the network, and the probability is found by studying the non-diagonal values in the distance matrix. The code below shows how to compute both (including a plot of the probability of randomly picking a path with a particular length or less).
Good luck!
Tore
11. Drahomir | March 3, 2016 at 8:20 pm
Drahomir | March 3, 2016 at 8:20 pm
Thank you very much Dr. Opsahl, really appreciating your kind help.
12. Drahomir | March 20, 2016 at 10:11 pm
Drahomir | March 20, 2016 at 10:11 pm
Hello again Dr. Opsahl
I’ve recently come about 2 uncertaineties. I’ve analyzed a graph consisting of 157 nodes. When i compute a matrix of weighted distances (shorthest paths) between nodes my matrix is 141×141 not 157×157 (as I understand it, this is mainly because of existing separated dyads (pairs of 2 nodes connected only to each other and disconnected from all other nodes in graph). There are 7 of these separated pairs in my graph. Could this be a reason for recieving 141×141 matrix and not the 157×157 matrix?
Secondly. If i’m looking at the specific value of distance in matrix lets say row = 28 and collumn = 31, is this value a distance between nodes (28-31) that were originaly inputed in the analyzed dataframe? In other words, is row number 28 representing node number 28 in original data?
Thanks for any advice,
With regards
Drahomír
13. Tore Opsahl | March 25, 2016 at 12:53 am
Tore Opsahl | March 25, 2016 at 12:53 am
Hi Drahomír,
Glad you are able to use tnet. To answer your questions in order:
1) The distance matrix is only from the main / giant component of the network. If you would like it for all nodes, you must specify gconly=FALSE in the distance_w-function.
2) The node ids of the rows and columns in the distance matrix when gconly=TRUE is saved as an attribute of the output from the distance_w-function. For example, if the output object is called dmat, the node list can be found by typing attributes(dmat)$nodes.
Good luck!
Tore
14. Drahomir | April 12, 2016 at 2:08 am
Drahomir | April 12, 2016 at 2:08 am
Thanks Dr. Opsahl
Unfortunately I’m still a bit confused.
Here’s my problem. Nodes in my graph represent animal names, produced in category/semantic fluency test. 2 most strongly connected nodes are “cat” and “dog”. I cannot think of any other shortest path between these two nodes than the direct one, as this is the path with highest weight in graph. Node “cat” was numericaly labeled as 1 and node “dog” as 2. When i lookup shorthest path between 1 and 2 in dmat matrix the value is 2.74 and this doesn’t make any sense to me.
I’m definitely missing something, would be glad for any further guidance.
15. Tore Opsahl | April 13, 2016 at 5:04 am
Tore Opsahl | April 13, 2016 at 5:04 am
Hi Drahomir,
Please send me an email with your data and code, and I will have a look at what is going on.
Tore
16. Drahomir | April 17, 2016 at 10:47 am
Drahomir | April 17, 2016 at 10:47 am
Hi, I’m really appreciating your willingness to help, but I’ve already figured it out. Error was in my data input, which didn’t include raw weights but already normalized, so they were normalized once again by the package.
17. Elio Bartos | April 1, 2016 at 2:33 pm
Elio Bartos | April 1, 2016 at 2:33 pm
Hello,
thanks for this post, I’m just getting started with SNA and tnet package.
1. Is there a way to calculate shortest distances like in example 1. (without normalisation) with tnet package?
2. Does measures like closeness use normalised shortest distances or just shortest distances?
Thank you!
18. Tore Opsahl | April 1, 2016 at 3:11 pm
Tore Opsahl | April 1, 2016 at 3:11 pm
Hi Elio,
1) The “normalization” of ties weights is a constant applied to all weights. You can undo this normalization with the mean time weight. Have a look at the function by typing distance_w without the (). You can then copy this code and comment out the normalization line.
2) The functions rely on the normalized distances. But it’s key to remember that the same paths are identified as the shortest ones with and without normalization.
Hope this helps!
Tore
19. Prem | July 14, 2016 at 11:29 am
Prem | July 14, 2016 at 11:29 am
Hello Tore
I am using tnet package for my weighted network analysis. I am little bit confuse for average path length of disconnected graphs. After breakdown my network divided into 10 part, where i am considering the sub graph have more than 5 nodes, most of the algorithm is to calculate the mean path length by considering the number of nodes from giant component only. But in a power grid network , each sub graph is a part of the network even if there is no link for a certain period of time, so i wish to consider all the nodes present in disconnected sub graph, and wants to calculated average path length based on total nodes, am i right or wrong?
20. Tore Opsahl | July 14, 2016 at 8:59 pm
Tore Opsahl | July 14, 2016 at 8:59 pm
Hi Prem,
Path lengths are infinite among nodes in disconnected component (i.e., no path exists between them). The average of a vector with an infinite number is infinite. If you’re looking to compute closeness, see https://toreopsahl.com/2010/03/20/closeness-centrality-in-networks-with-disconnected-components/
Best,
Tore
21. Prem | July 16, 2016 at 12:19 pm
Prem | July 16, 2016 at 12:19 pm
thank you Tore
22. Gab (@red_peque) | October 23, 2016 at 9:59 pm
Gab (@red_peque) | October 23, 2016 at 9:59 pm
Hello Dr. Opsahl!
First of all, many thanks for tnet and all the information in your blog.
Sorry, I am having a problem when trying to get distances in a weighted directed network. I am inputting data that contains 304 nodes and 710 edges. 300 nodes are connected to the giant component and only 4 are not. I was expecting to get a matrix of 300*300 (or 304*304) but I am only getting 66*66 (when using “gconly=TRUE”). I tried to write the “distance_w” function with “gconly=FALSE” but I get a message saying that R has encountered a fatal error and it needs to shut down. Any clue of what I might be doing wrong? (My weights take the values of 1, 2, 2.5 and 3 – So not sure if the ‘.5’ is creating an issue)
Many thanks!
Gabriela
23. Tore Opsahl | October 26, 2016 at 5:12 pm
Tore Opsahl | October 26, 2016 at 5:12 pm
Hi Gabriela,
Glad you are finding the information useful.
I am not entirely sure what is happening in your case. Please drop me a note with your network and code. Then I can better help you.
The 0.5 weight should not be an issue.
Best,
Tore
24. Mehrzad | April 6, 2017 at 3:18 pm
Mehrzad | April 6, 2017 at 3:18 pm
Hi, thanks for your good article. I just have question. Suppose there is an tie with weight 2 between two nodes A, B. However, there is another path from A to B like this: ACDEB with weight 3 for each tie [(A,C)=3, (C,D)=3, (D,E)=3, (E,B)=3)]. As you said the strongest path would be better path. So the algorithm must return second path as the solution but it does not, because the first path will have lower value 1/2 while the second one has the value (1/3+1/3+1/3+1/3=4/3). I was confused by this. I will appreciate you if give me an answer.
Thanks
Mehrzad
25. Tore Opsahl | April 10, 2017 at 5:09 pm
Tore Opsahl | April 10, 2017 at 5:09 pm
Hi Mehrzad,
Glad the post is helpful.
The distance is the sum of inverse tie weights, so an indirect path consisting of two ties with weights of 10 would be preferred over a direct path with a tie weight of less than 5 as 1/10 + 1/10 < 1/4.
Best,
Tore
26. Hillary | January 10, 2018 at 5:08 pm
Hillary | January 10, 2018 at 5:08 pm
Dear Dr. Opsahl,
Thank you so much for these posts! I’ve found them very helpful as I am doing research for my senior thesis. I was hoping to calculate in-closeness and out-closeness centrality and was wondering if there was a way, either in tnet or some other package, that you were aware of for calculating these measures? I’ve been reading a paper by De Benedictis et al., called “Network Analysis of World Trade using the BACI-CEPII Dataset” and they were able to implement it in the tnet library. I’d appreciate any ideas you may have.
Thanks,
Hellary
27. Tore Opsahl | January 10, 2018 at 5:20 pm
Tore Opsahl | January 10, 2018 at 5:20 pm
Hi Hillary,
Have a look at this page: https://toreopsahl.com/tnet/weighted-networks/node-centrality/
Specifically, you can compute the standard closeness measure (ie inverted sum of distances to all other nodes) by running closeness_w-function on the network. If you want the reverse closeness measure (ie inverted sum of distances from all other nodes), then you can precompute the distance matrix using distance_w, transpose the matrix, and use this matrix in the closeness_w-function (precomp.dist-parameter).
Hope this helps,
Tore
28. Hellary Zhang | January 13, 2018 at 10:56 pm
Hellary Zhang | January 13, 2018 at 10:56 pm
I see. Thank you for your help!
So just to make sure I understand, for a weighted and directed network, closeness_w(net) would calculate out-closeness, but if I wanted to find in-closeness, I could do:
trans_distance_mat <- t(distance_w(el_1995_final, gconly=FALSE))
w_incloseness_1995 <- data.frame(closeness_w(el_1995_final, gconly=FALSE, precomp.dist = trans_distance_mat))
Additionally, I think I am still a bit confused as to how closeness_w uses distance_w in its calculations. I tried calculating closeness_w by hand for the example network given here (https://toreopsahl.com/tnet/weighted-networks/shortest-paths/) but am not getting the same numbers that are computed in R. Would you mind sharing how exactly closeness_w is calculated?
I calculated closeness by hand by using the equations in page 30 of this paper: http://www.cepii.fr/PDF_PUB/wp/2013/wp2013-24.pdf.
Thanks so much again,
Hillary
29. Tore Opsahl | January 17, 2018 at 3:35 am
Tore Opsahl | January 17, 2018 at 3:35 am
Hi Hillary,
A couple of points:
1) closeness can only be computed for the giant component; when you specify gconly=FALSE, an alternative approach is taken (see https://toreopsahl.com/2010/03/20/closeness-centrality-in-networks-with-disconnected-components/)
2) to inspect the code behind the functions, simply type them without the () at the end
3) the equations in that paper normalized the coefficients; the functions in tnet do not do this
Hope this helps!
Tore
30. Alex | January 26, 2018 at 3:45 pm
Alex | January 26, 2018 at 3:45 pm
Hi Tore,
first of all I’d like to also thank you for your very helpful and interesting approaches on SNA.
If have one question and I’d very grateful if you could help me in this case.
I applied the distance_w function on two different network as described above. Unfortunately, in one case the result was NaN but I was not able to find out why. Both networks are significantly different in terms of their size. The network with the result NaN has very strong betweeness nodes. Could that be a reason for the result? Or is there a way how to interpret NaN as a result in this specific case?
Best regard and thanks a lot in advance,
Alex
31. Tore Opsahl | January 29, 2018 at 2:41 am
Tore Opsahl | January 29, 2018 at 2:41 am
Hi Alex,
The distance_w-function considers the directionality of ties. If the network is not fully connected, you might get NaNs. Please email with your data and code, and I will look into it.
Best,
Tore
32. Katy | August 5, 2022 at 3:43 am
Katy | August 5, 2022 at 3:43 am
Dear Dr. Opsahl,
Thank you so much for your blog’s useful information! I’ve found them very helpful as I am researching social network analysis.
When trying to get the distances in a weighted directed network, I have a problem. In my network, the inverse tie weights are different, and I want the path distance to be different when two vertices are “from” and “to,” respectively. For example, the weight from A to B is 2, but from B to A will be 1. However, as you mentioned, distance is the sum of inverse tie weights. So I wonder if there is any way to separately calculate the inverse tie weights to get the path distance.
The tutorials and functions I found online currently cannot address this situation, and I would like to know if you have any suggestions to tackle this problem.
Thanks so much for your help,
Katy
33. Tore Opsahl | August 9, 2022 at 6:44 am
Tore Opsahl | August 9, 2022 at 6:44 am
Hi Katy,
The distances do incorporate direction of ties and their weight. The distance_w-function computes the distance matrix where the cell i,j will be the distance from node i to node j. The upper triangle and lower triangle will match for undirected networks, but not for directed networks (i.e., cell i,j is not the same as j,i).
In your example, the distance matrix will be
A B
A 0 0.5
B 1 0
Hope this was helpful,
Tore