Article: Node centrality in weighted networks: Generalizing degree and shortest paths
April 21, 2010 at 10:37 am 95 comments
A paper called “Node centrality in weighted networks: Generalizing degree and shortest paths” that I have co-authored with Filip Agneessens and John Skvoretz will be published in Social Networks. Unfortunately, the copyright agreement prevents me from uploading a pdf of the published paper to this blog. However, if you have access to Social Networks, you can download the paper directly. Otherwise, a preprint with the exact same text is available.
Abstract
Ties often have a strength naturally associated with them that differentiate them from each other. Tie strength has been operationalized as weights. A few network measures have been proposed for weighted networks, including three common measures of node centrality: degree, closeness, and betweenness. However, these generalizations have solely focused on tie weights, and not on the number of ties, which was the central component of the original measures. This paper proposes generalizations that combine both these aspects. We illustrate the benefits of this approach by applying one of them to Freeman’s EIES dataset.
Motivation

Ego networks of Phipps Arabie (A), John Boyd (B), and Maureen Hallinan (C) from Freeman's third EIES network. The width of a tie corresponds to the number of messages sent from the focal node to their contacts. Adopted from the paper.
The three measures have already been generalised to weighted networks. Barrat et al. (2004) generalised degree to weighted networks by taking the sum of weights instead of the number ties, while Newman (2001) and Brandes (2001) utilised Dijkstra’s (1959) algorithm of shortest paths for generalising closeness and betweenness to weighted networks, respectiviely. Dijkstra’s algorithm defined the length of paths as the sum of cost (e.g., time in GPS calculations), which is generally only defined as the sum of the inversed tie weights. All these generalisations fail to take into account the main feature of the original measures formalised by Freeman (1978): the number of ties.
This limitation is highlighted for degree centrality by the three ego networks from Freeman’s third EIES network. The three nodes have roughly sent the same amount of messages; however, to a quite different number of others. If Freeman’s (1978) original measure was applied, the centrality score of the node in panel A is almost five times as high as the node in panel C attains. However, when using Barrat et al.’s generalisation, they get roughly the same score.
This articles proposes a new generation of node centrality measures for weighted networks. The second generation of measures takes into consideration both the weight of ties and the number of ties. The relative importance of these two aspects are controlled by a tuning parameter.
Want to test it with your data?
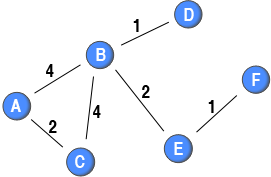 The degree_w, closeness_w, and betweenness_w-functions in tnet allows you to calculate the binary, weighted, and the measures that combine these two aspects on your own dataset.
The degree_w, closeness_w, and betweenness_w-functions in tnet allows you to calculate the binary, weighted, and the measures that combine these two aspects on your own dataset.
For example, to calculate second generation node centrality measures (alpha = 0.5) on the sample network above, you can run the code below in R. The degree function easily calculates the binary and first generation measures as well; however, this is not the case for the closeness and betweenness-functions. If you would like the binary version, you can either use the dichotomise function or set alpha=0. If you would like the first generation weighted measures, you can set alpha=1 (default value).
# Load tnet
library(tnet)
# Load network
net <- cbind(
i=c(1,1,2,2,2,2,3,3,4,5,5,6),
j=c(2,3,1,3,4,5,1,2,2,2,6,5),
w=c(4,2,4,4,1,2,2,4,1,2,1,1))
# Calculate degree centrality (note that alpha is included in the list of measures)
degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5)
# Calculate closeness centrality
closeness_w(net, alpha=0.5)
# Calculate betweenness centrality
betweenness_w(net, alpha=0.5)
To test it on Freeman’s third EIES network from the datasets-page and recreate Table 3 of the paper, you can do the following:
# Load tnet
library(tnet)
# Load network
data(Freemans.EIES)
net <- Freemans.EIES.net.3.n32
# Calculate measures
tmp <- data.frame(
Freemans.EIES.node.Name.n32,
degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5),
degree_w(net, measure="alpha", alpha=1.5)[,"alpha"], stringsAsFactors=FALSE)
dimnames(tmp )[[2]] <- c("name", "node", "a00", "a10", "a05", "a15")
tmp <- tmp[,c("name","a00","a05","a10","a15")]
# Merge names and order table
out <- data.frame(
seq.int(nrow(tmp)),
tmp[order(-tmp[,"a00"], -tmp[,"a10"]),c("name", "a00")],
tmp[order(-tmp[,"a05"], -tmp[,"a10"]),c("name", "a05")],
tmp[order(-tmp[,"a10"], -tmp[,"a10"]),c("name", "a10")],
tmp[order(-tmp[,"a15"], -tmp[,"a10"]),c("name", "a15")])
dimnames(out)[[2]] <- c("Rank",
"a00.name","a00",
"a05.name","a05",
"a10.name","a10",
"a15.name","a15")
# Display table
out
References
Brandes, U., 2001. A Faster Algorithm for Betweenness Centrality. Journal of Mathematical Sociology 25, 163-177.
Dijkstra, E. W., 1959. A note on two problems in connexion with graphs. Numerische Mathematik 1, 269-271.
Freeman, L. C., 1978. Centrality in social networks: Conceptual clarification. Social Networks 1, 215-239.
Newman, M. E. J., 2001. Scientific collaboration networks. II. Shortest paths, weighted networks, and centrality. Physical Review E 64, 016132.
Opsahl, T., Agneessens, F., Skvoretz, J. (2010). Node centrality in weighted networks: Generalizing degree and shortest paths. Social Networks 32, 245-251.
Entry filed under: Articles. Tags: actors, arcs, betweenness, centrality, closeness, complex networks, degree, directed networks, edges, graphs, gregariousness, hubs, Links, local, network, nodes, popularity, reinforcement, shortest distance, shortest path, social network analysis, strength of nodes, strength of ties, ties, undirected networks, valued networks, vertices, weighted networks.
 RSS feed
RSS feed
1. questavita | May 2, 2010 at 10:54 am
questavita | May 2, 2010 at 10:54 am
Thank you for your very good works.
—
Jacopo, from Rome, Italy.
2. Marco | May 13, 2010 at 10:42 am
Marco | May 13, 2010 at 10:42 am
Hi Tore,
just one quick question (I’m pretty new in this stuff): with cbind what are you passing to the programme exactly? I dont understand why there are 3 parameters (i,j,w) with 12 elements..
Marco – Italy
3. Tore Opsahl | May 13, 2010 at 12:53 pm
Tore Opsahl | May 13, 2010 at 12:53 pm
Hi Marco,
With cbind you combine columns. You can get the help file for each function in R by typing ?cbind
The command:
net <- cbind(
i=c(1,1,2,2,2,2,3,3,4,5,5,6),
j=c(2,3,1,3,4,5,1,2,2,2,6,5),
w=c(4,2,4,4,1,2,2,4,1,2,1,1))
I create an object called net that is composed of three columns (i, j, and w), which contain the values given in inside the vector-combination function, c.
Hope this helps,
Tore
4. Marco | May 17, 2010 at 7:25 am
Marco | May 17, 2010 at 7:25 am
Hi Tore, thanks for your answer. Still, I got a doubt: the values in the columns, what do they correspond to? I mean, why is the vector i made of 1,1,2,2,2,2 etc? Because, looking to the net represented by the picture (the net made by 6 nodes, from A to F) I see other weigths, so I am pretty lost :)
Looking forward for your reply,
Marco
5. Marco | May 17, 2010 at 9:35 am
Marco | May 17, 2010 at 9:35 am
Tore, I’ve just understood the values..but I’d have another quick question: what’s alpha?
Sorry for disturbing you:)
Marco
6. Tore Opsahl | May 17, 2010 at 11:27 pm
Tore Opsahl | May 17, 2010 at 11:27 pm
Glad that you figured it out! Have a look at the paper for the alpha parameter!
7. Thor Sigfusson | June 14, 2010 at 3:47 pm
Thor Sigfusson | June 14, 2010 at 3:47 pm
Tore,
Thanks for your help. I am still figuring out out to work on weighted data.
What do the numbers in the columns represent ? (1,1,2,2,3,3 etc.
I have a data where names of individuals in the network (the nodes) are 1-300. I interviewed 20 entrepreneurs who have numbers from 1-20. Each mentioned around 15-20 connections.
Person 1 has strong ties with 5,12, 19,21 and weak ties with 12 others.
I am stryggling with setting this up in the correct manner.
Best regards and thanks again for a great initiative,
Thor
8. Tore Opsahl | June 15, 2010 at 6:01 am
Tore Opsahl | June 15, 2010 at 6:01 am
Hi Thor,
tnet cannot -so far- handle node labels, so they are simply the id numbers of the nodes.
The best way to enter larger networks is to load an edgelist file. You can do this by arranging your data in a txt file like this:
1 5 5
1 12 5
1 19 5
1 21 5
1 4 1
….
Then write in R: net <- read.table("filename.txt")
Hope this helps,
Tore
9. Thor Sigfusson | June 24, 2010 at 11:30 am
Thor Sigfusson | June 24, 2010 at 11:30 am
Dear Tore,
It worked. Thanks a lot.
Thor
10. Marco | June 30, 2010 at 10:18 am
Marco | June 30, 2010 at 10:18 am
Hi Tore,
got a quick question, maybe you can help me: I’m calculating the EigenValues of a Matrix, but R provides me only ordered values..is there a way to have unordered results?
Thnaks in advance
Marco
11. Ben | July 24, 2010 at 10:19 am
Ben | July 24, 2010 at 10:19 am
Hi Tore,
What kind of software do you use for drawing your graph pictures?
For example, this one:
http://thetore.files.wordpress.com/2008/12/fig1.png?w=271&h=177
Thanks,
Ben
12. Tore Opsahl | July 24, 2010 at 4:15 pm
Tore Opsahl | July 24, 2010 at 4:15 pm
Hi Ben,
I have actually made those in illustrator / photoshop. Not recommended, but that method enables you to make the graphs exactly as you want.
Best,
Tore
13. Mauricio | August 10, 2010 at 2:19 pm
Mauricio | August 10, 2010 at 2:19 pm
Hi Tore,
Thanks for your very good work.
I’m trying to calculate the degree_w for my data, but it seems that the output is always equivalent when alpha is set as 1. Even for the sample network in this post, I got the same results when varying the alpha parameter…
Please, could you tell me what is my mistake here?
With many thanks
Mauricio
> degree_w(net,measure=c(“degree”,”output”), type=”out”, alpha=1)
node degree output
[1,] 1 2 6
[2,] 2 4 11
[3,] 3 2 6
[4,] 4 1 1
[5,] 5 2 3
[6,] 6 1 1
> degree_w(net,measure=c(“degree”,”output”), type=”out”, alpha=0.5)
node degree output
[1,] 1 2 6
[2,] 2 4 11
[3,] 3 2 6
[4,] 4 1 1
[5,] 5 2 3
[6,] 6 1 1
14. Tore Opsahl | August 11, 2010 at 12:37 am
Tore Opsahl | August 11, 2010 at 12:37 am
Mauricio,
You need to include alpha in the measure-parameter.
Tore
#Load tnet and network library(tnet) net <- cbind( i=c(1,1,2,2,2,2,3,3,4,5,5,6), j=c(2,3,1,3,4,5,1,2,2,2,6,5), w=c(4,2,4,4,1,2,2,4,1,2,1,1)) # Run function degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5) # Output node degree output alpha [1,] 1 2 6 3.464102 [2,] 2 4 11 6.633250 [3,] 3 2 6 3.464102 [4,] 4 1 1 1.000000 [5,] 5 2 3 2.449490 [6,] 6 1 1 1.00000015. Mauricio | August 11, 2010 at 1:23 pm
Mauricio | August 11, 2010 at 1:23 pm
Hi Tore,
Thank you for your reply, it’s fixed my problem!!
I didn’t notice this detail in the post. I was following the help(degree_w) from the tnet package, and I got confused.
With many thanks,
M
16. Nitesh | August 18, 2010 at 2:37 pm
Nitesh | August 18, 2010 at 2:37 pm
Hi Tore,
Great work!!
Just having problem in installing tnet package in R 2.11.1.
The error I am getting is
Loading required package: igraph
Error : .onLoad failed in loadNamespace() for ‘igraph’, details:
call: inDL(x, as.logical(local), as.logical(now), …)
error: unable to load shared library ‘C:/PROGRA~1/R/R-211~1.1/library/igraph/libs/igraph.dll’:
LoadLibrary failure: The specified module could not be found.
Error: package ‘igraph’ could not be loaded
In the pop window, it is showing that iconv.dll file was not found..
Can you help me please??
17. Tore Opsahl | August 18, 2010 at 2:52 pm
Tore Opsahl | August 18, 2010 at 2:52 pm
Nitesh,
Thanks for taking an interest in tnet. This is an issue with the igraph-package that tnet depends on. You can fix it by copying the iconv.dll from a previous version of igraph (or email me). I have sent them a bug report – hopefully they will have it sorted soon.
Best,
Tore
18. Nitesh | August 18, 2010 at 3:56 pm
Nitesh | August 18, 2010 at 3:56 pm
Many thanks Tore,
Nitesh
19. Tore Opsahl | August 19, 2010 at 12:28 pm
Tore Opsahl | August 19, 2010 at 12:28 pm
Nitesh,
The igraph-guys (Gabor Csardi) have just submitted a new version of their R-package that should solve this issue. As soon as it has been approved by CRAN and disseminated across the servers, you should be able to get it by writing update.packages() in R.
Tore
20. Nitesh | August 19, 2010 at 12:35 pm
Nitesh | August 19, 2010 at 12:35 pm
Hi Tore,
Thanks for your response. As you mentioned before, by copying the iconv.dll file in igraph-libs, it worked.
Can you suggest me something about cliques in weighted graphs. I have seen your related to node centrality and clustering coefficient.
Just wondering, if you have any ideas on finding cliques in weighted graph.
Regard
Nitesh
21. Tore Opsahl | August 19, 2010 at 1:42 pm
Tore Opsahl | August 19, 2010 at 1:42 pm
Nitesh,
You can look at my paper called Clustering in Weighted Networks for determining the level of clustering / cliqueness in a network. But if you would like to find the specific cliques or communities, I would suggest you contact some of the community detection guys. Also, you might want to have a look at the book Generalized Blockmodeling by Dorian, Batagelj, and Ferligoj.
Tore
22. minjung | August 24, 2010 at 12:55 pm
minjung | August 24, 2010 at 12:55 pm
Hi tore!
Thanks for sharing ur great works. It’s really helpful for me to understand
the concepts about centrality.
I just wondering u can give me some advice.
Im working on a project that analyzing the customers of mobile company and doing SNS. That is, i have to find the community among the customers and define the leader, who is influential to other customers.
So i detect the community(cluster) and i got the degree centrality, closeness centrality, between centrality of each nodes within a cluster.
And NOW, i have to assign roles to each node; leader, sub-leader,follower and outliers.
Do u think i can just assign roles based on three centrality? for example,
make the one with big centralites as a leader. Centrality can be the measure of influence within the community?
Thanks in advance,
MJ
23. Tore Opsahl | August 24, 2010 at 7:03 pm
Tore Opsahl | August 24, 2010 at 7:03 pm
Hi MJ,
I am not aware of any general literature on this topic. You could have a look at the Fernandez and Gould-paper that does something similar with brokerage measures. They define – based on directionality, which your data should have – various roles based on ego networks.
Best,
Tore
24. Vícktor | October 7, 2010 at 7:15 pm
Vícktor | October 7, 2010 at 7:15 pm
Hello,
Hi,
I have a doubt. In his article states that could make a regression analysis to find an optimal level of alpha. For example, if I have a binary variable to explain, how you might proceed in the analysis?. You could provide guidance for compare different and found and optimal alpha. I use SPSS for analysis.
Thanks!
Victor Hugo
25. Tore Opsahl | October 8, 2010 at 12:16 am
Tore Opsahl | October 8, 2010 at 12:16 am
Victor,
Thank you for taking an interest in my work, and reading the future research section carefully!
The “optimal” (read: what high performers have)-level of the centrality measures could be probed by using a performance variable as the dependent variable and a centrality variable as an independent variable along with controls. If you ran multiple regressions each with a different alpha for the centrality parameter (e.g., 0, 0.1, 0.2, 0.3 etc), then if you could plot the attained z-scores of the centrality variable (y-axis) against the alpha (x-axis). This should give you an inverse u-shaped curve. The optimal-level is where the maxium z-score is attained.
Hope this helps,
Tore
26. Viktor | October 9, 2010 at 4:25 pm
Viktor | October 9, 2010 at 4:25 pm
Hi Tore!
Thanks. When I have the analysis, I will contact you again. Maybe, I made a good discovery with your idex. Let me try!!!
Sincerelly,
Vicktor
27. Rainbow Socks | January 28, 2011 at 1:17 pm
Rainbow Socks | January 28, 2011 at 1:17 pm
Hi Tore,
Great site, and great software :)
One question regarding degree_w and closeness_w. The output I get is an array, however I am interested in the overall degree(strength) centralization of the weighted network, not for each individual node. Is there a way to do this with tnet?
Best,
Kirsten
28. Tore Opsahl | January 28, 2011 at 4:48 pm
Tore Opsahl | January 28, 2011 at 4:48 pm
Hi Kirsten,
Thank you for using the site and tnet!
Normalisation of node centrality scores is, to my opinion, adding a bias to the data instead of removing one. In fact, I have stayed clear of standardising the measures due to what I believe was misleading in the original measures, let alone generalised ones. My main concern with the original ways of standardising/normalising node centrality measures (i.e., n-1) is that these scale linearly with the number of nodes. Specifically, I believe that none of the main three node centrality measures scales linearly. First, it has been argued that the average degree in networks does not change as a network grows. Hence, no scaling (i.e., use the average degree to compare networks). Second, closeness centrality is based on shortest distances. In small world networks, shortest distances does not scale linearly with the number of nodes, but rather logarithmically (i.e., divide farness scores by log(N) if your network is a “small world”). Third, betweenness is based on n*(n-1) shortest paths, so it could be argued that it scales n-squared. Given these issues with the original measures, I have not given much thought/effort to normalise the generalised ones. Let me know if you figure out a way of doing it!
Best,
Tore
29. Rainbow Socks | January 28, 2011 at 9:51 pm
Rainbow Socks | January 28, 2011 at 9:51 pm
Hi Tore,
Thank you for the quick response. I will have to think about this… :)
Just to make sure that I get your other function right, – the clustering_w “gm” would be a transitivity measure for the overall weighted graph, right?
Best,
Kirsten
30. Tore Opsahl | January 29, 2011 at 12:16 am
Tore Opsahl | January 29, 2011 at 12:16 am
Kirsten,
All the clustering_w-function is the global clustering coefficient, while the clustering_w_local-function is the local clustering coefficient that produces a score for each node (see Barrat et al., 2004). These functions are just different aggregations of the triplets in the network with the global one aggregating all triplets and the local one being an intermediate step.
The measure-parameter (“am” “gm” “ma” “mi” “bi”) controls how triplets are valued. This could, for example, be the regular mean (arithmetic mean; “am”) of the two tie weighs that make up the triplet, or the geometric mean (“gm”). The main difference between these two is that the geometric mean discounts the tie weight if there is variation (e.g., tie weights of 2 and 2 would be am=2 and gm=2, and tie weights 3 and 1 would be am=2 and gm=sqrt(3)~1.73.
Hope this helps,
Tore
31. Rainbow Socks | January 29, 2011 at 9:27 am
Rainbow Socks | January 29, 2011 at 9:27 am
That helps, thanks.
One more quesiton… concerning degree_w, is there a way to calculate the degree_w per node using the inverse weights with tnet?
Best,
Kirsten
32. Rainbow Socks | January 29, 2011 at 9:55 am
Rainbow Socks | January 29, 2011 at 9:55 am
Hi again Tore,
Sorry for ‘spamming’, I forgot to ask another question that puzzels me: I am using closeness_w, but if I dont use weights (set them all to 1, just to check), I do not get the same result for node closeness as when using closeness() in an unweighted network. Why is this? Am I doing something wrong?
Kirsten
33. Tore Opsahl | January 30, 2011 at 3:10 pm
Tore Opsahl | January 30, 2011 at 3:10 pm
Kirsten,
I am uncertain what you are referring to when you are using inverse weights in the degree_w-function, and also what closeness() is. Please send me an email with the commands and data you are using.
Best,
Tore
34. xingqin | March 16, 2011 at 2:31 am
xingqin | March 16, 2011 at 2:31 am
Hi, Tore, Thanks for your hard work, it is easy to use your code now, just simply add \alpha. One question, do you know are there any other centrality methods handing weighted networks yet? Eigenvector is also a centrality method, but it is dealing with unweighted graphs, Does the same idea work for weighted ones?
Thanks a lot.
xingqin
35. Victor | March 19, 2011 at 2:50 pm
Victor | March 19, 2011 at 2:50 pm
Hi Tore,
I want to calculate degree_w. I know that for out degree I need to write..
degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5)But, I dont know how to write the instruction for calculate in degree.
degree_w(net, measure=c("degree", "in", "alpha"), alpha=0.5).It’s correct or not the second one.
May I have your help.
Have a good day,
Sincerely, Victor
36. Tore Opsahl | March 19, 2011 at 4:35 pm
Tore Opsahl | March 19, 2011 at 4:35 pm
Hi Victor,
The degree_w-function uses the outgoing ties by default. To calculate the measures for the incoming ties, you should define type=”in”.
degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5, type="in")For an undirected network, the type parameter is irrelevant as the outbound and incoming ties are identical. For example:
> # Load tnet > library(tnet) > # Load network > net <- cbind( > i=c(1,1,2,2,2,2,3,3,4,5,5,6), > j=c(2,3,1,3,4,5,1,2,2,2,6,5), > w=c(4,2,4,4,1,2,2,4,1,2,1,1)) > > # Calculate degree centrality > degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5) node degree output alpha [1,] 1 2 6 3.464102 [2,] 2 4 11 6.633250 [3,] 3 2 6 3.464102 [4,] 4 1 1 1.000000 [5,] 5 2 3 2.449490 [6,] 6 1 1 1.000000 > > degree_w(net, measure=c("degree", "output", "alpha"), alpha=0.5, type="in") node degree output alpha [1,] 1 2 6 3.464102 [2,] 2 4 11 6.633250 [3,] 3 2 6 3.464102 [4,] 4 1 1 1.000000 [5,] 5 2 3 2.449490 [6,] 6 1 1 1.000000Best,
Tore
37. Nicola | May 10, 2011 at 10:06 am
Nicola | May 10, 2011 at 10:06 am
To Tore,
Forgive me if this is an ignorant question, but I was just wondering what exactly the values in the output meant in terms of the values given for “degree”, “output” and “alpha”?
Thanks for your help.
38. Tore Opsahl | May 10, 2011 at 12:01 pm
Tore Opsahl | May 10, 2011 at 12:01 pm
Nicola,
The values are:
-degree: number of ties
-output: sum of tie weights (aka node strength)
-alpha: the second generation weighted measure that combines degree and output, see the paper.
You can also find this by typing ?degree_w in R.
Best,
Tore
39. Victor | May 17, 2011 at 11:23 pm
Victor | May 17, 2011 at 11:23 pm
Hi Tore,
I have a question again,
I want to study how alpha value helps predict a continuous variable.
My database capture a communication network and I want to find what value of alpha helps to explain a continuous variable (for example, to predict the number of words used by each node using weighted out degree). Ridge-regression method can be a good choice ?
Sincerely, Victor
40. Tore Opsahl | May 18, 2011 at 11:12 am
Tore Opsahl | May 18, 2011 at 11:12 am
Hi Victor,
Your data sounds ideal for finding the “optimal” value of alpha. My understanding of ridge models are limited, but here is a suggestion of how it can be done using OLS. Note that the example data is not ideal as there are only 32 observations, the dependent variable is not normally distributed, and there are no control variables etc, but it shows how it can be implemented.
# Load tnet and data library(tnet) data(Freemans.EIES) # Define output data frame out <- data.frame(alpha=seq(from=0, to=2, by=0.1), y=NaN) # Get independent variables for(i in 1:nrow(out)) { # Compute centrality measure tmp <- degree_w(net=Freemans.EIES.net.3.n32, measure="alpha", alpha=out[i,"alpha"]) # Extract just centrality scores tmp <- tmp[,"alpha"] # Regress (add control variables etc) reg <- lm(Freemans.EIES.node.Citations.n32 ~ tmp) # Instead of a nice way of doing it: Extract the t-statistic tmp <- eval(parse(text=as.character(summary(reg))[4])) tmp <- matrix(data=tmp, nrow=(length(tmp)/4), ncol=4) tmp <- abs(tmp[nrow(tmp),3]) out[i,"y"] <- tmp } # Plot alpha values and the corresponding significance obtained plot(out, type="b", ylab="Significance of centrality measure")The last command produces the following plot, which suggests that for this limited data, the optimal point is close to 0. Please do not base any conclusions on this data due to the limitations listed above.
Hope this helps,
Tore
41. Nicola | May 18, 2011 at 11:25 am
Nicola | May 18, 2011 at 11:25 am
Hi Tore,
I read this post with interest … would the same work if the dependent variable was a factorial measure? In my case it would be animal disease status as the dependent variable – either positive or negative, and seeing which value of alpha was most appropriate to infer whether the number of animals you are connected to (an alpha value closer to 0) or the weighting – the amount of time that you spend in contact with those animals (an alpha value closer to 1) was more important in terms of predicting whether individuals would be infected or not?
Thanks for your help,
Nicola
42. Tore Opsahl | May 18, 2011 at 9:39 pm
Tore Opsahl | May 18, 2011 at 9:39 pm
Nicola,
The above code should work for other cases; however, you should change the line starting with “reg <- lm(" to a regression function that suits your dependent variable.
Let me know if you have any issues,
Tore
43. Victor | May 18, 2011 at 8:02 pm
Victor | May 18, 2011 at 8:02 pm
Thanks! Tore.
You are the best!
44. Nicola | July 31, 2011 at 11:01 am
Nicola | July 31, 2011 at 11:01 am
Hi Tore,
I was wondering if you could help me with an error message that i’m getting? I’m doing some simple analysis with a weighted network and have calculated degree and closeness values fine, but the betweenness_w function is throwing up an error message:
> warnings()
Warning messages:
1: In get.shortest.paths(g, from = i, to = V(g)[V(g) > i]) :
At structural_properties.c:4277 :Couldn’t reach some vertices
I’m using the same edgelist for this as for the degree and closeness values that worked fine.
Thanks for any help that you can give me!
Nicola
45. Tore Opsahl | July 31, 2011 at 8:17 pm
Tore Opsahl | July 31, 2011 at 8:17 pm
Hi Nicola,
Thanks for using tnet. Could you send me a copy of your data and code that you are using?
Best,
Tore
46. Tore Opsahl | August 1, 2011 at 4:11 pm
Tore Opsahl | August 1, 2011 at 4:11 pm
Hi Nicola,
Thanks for your data and code.
It seems that you are getting the warnings because you have disconnected components. All the components are fully connected except for the ones with node 31 and 42. In these two components, the two nodes sits between others, and hence gets a betweenness score when the alpha is set to 0.
Hope this helps,
Tore
47. Tara | November 27, 2012 at 3:32 pm
Tara | November 27, 2012 at 3:32 pm
Hi Tore,
IS it possible to calculate centrality for valued graphs with ucinet ?if yes should i dichtomize the data or not before starting calculation.
Tara
48. Tore Opsahl | November 27, 2012 at 3:44 pm
Tore Opsahl | November 27, 2012 at 3:44 pm
Hi Tara,
I am not entirely sure whether you can do it in UCINET. You can do it in tnet: https://toreopsahl.com/tnet/
Best,
Tore
49. Mathias | February 27, 2013 at 10:41 am
Mathias | February 27, 2013 at 10:41 am
Hi Tore,
Glad that I found your work. I am just wondering whether tnet can be used to derive the centrality of directed weighted network (i.e indegree & outdegree centrality of weighted network)?
Thanks.
Best regards,
Mathias
50. Tore Opsahl | February 27, 2013 at 2:10 pm
Tore Opsahl | February 27, 2013 at 2:10 pm
Hi Mathias,
Great! tnet is created around directed networks, and the degree_w-function calculates the out-degree binary scores, weighted scores, and the weighted scores from Node centrality in weighted networks: Generalizing degree and shortest paths (Social Networks 2010, 245-251). If you would like the corresponding in-degree scores, set the parameter type = “in”.
Best,
Tore
51. Michael Tsikerdekis | April 23, 2013 at 7:27 am
Michael Tsikerdekis | April 23, 2013 at 7:27 am
Hi Tore,
great work! It’s good to see that someone expanding the domain and methods.
I am doing comparative work across many networks. For this reason individual node centrality is not useful but what I need is to obtain the overall graph centrality. Is it appropriate to use the centralization formula used for the non weighted networks or perhaps obtain the mean of alpha values and compare this value across my various networks? Or should abandon the weighted approach all together and calculate non weighted normalization scores?
Best,
Michael
52. Tore Opsahl | April 23, 2013 at 3:57 pm
Tore Opsahl | April 23, 2013 at 3:57 pm
Hi Michael,
Thanks!
Normalization is not a straight forward process due to scaling issues with the number of nodes (see comment #28), and with weighted networks, you should also consider the distribution of weights. Given that the distributions are rarely normally distributed and networks are variously sparse, I have yet to come up with a good way to normalize networks. My recommendation is not to give up weighted network analysis for a binary one, but more to give up the more intricate normalization procedures and -perhaps- only use average scores.
If you come up with a better way, let me know!
Tore
53. Michael Tsikerdekis | May 3, 2013 at 12:04 pm
Michael Tsikerdekis | May 3, 2013 at 12:04 pm
Thank you Tore for your reply. I do agree that the weight matters alot and If I were to drop it then I would be missing important information. I could probably use weighted and not weighted just for comparison with the weighted using the mean scores perhaps.
Michael
54. Leila | May 21, 2013 at 7:31 pm
Leila | May 21, 2013 at 7:31 pm
Hi Tore,
Again, thank you for your research. I use it in the field of international economics and finance. Yet, I have a question. I would like to use the closeness centrality measure for directed network. Besides, my matrix of data is not squared (74 columns, 180 lines). When I add “in” in the program the argument is refused. For instance, closeness_w(net, alpha=0.5, type”in”) is not valid. I read the paper you wrote with Agneessens and Skvoretz. You indicate that for directed network, we need to add a constraint (“a path from one node to another can only follow the direction of present ties”). I don’t really understand, hence I have no idea how to transform my data to fill this constraint. Can you help me please ?
Thank you very much for your help Tore,
Kindest regards
Leila
55. Tore Opsahl | May 22, 2013 at 10:51 am
Tore Opsahl | May 22, 2013 at 10:51 am
Hi Leila,
Great that you are finding it useful. I am not entirely sure what kind of data that you have. If you send me an email with the network and data that you, then I can have a look at it.
Best,
Tore
56. Rui | August 31, 2013 at 8:38 pm
Rui | August 31, 2013 at 8:38 pm
Hi, Tore,
It is Rui again. I am reading you paper “Node centrality in weighted networks: Generalizing degree and shortest paths”. There are few questions about betweenness centrality in your paper. One is that in equation (6b), it seems that betweenness should be the summation of g_kj(i)/g_jk over j and k, doesn’t it? Another one is what is the expression of g^w\alpha_jk in equation (10)?
Thank you very much
Best
Rui
57. Tore Opsahl | September 3, 2013 at 3:06 am
Tore Opsahl | September 3, 2013 at 3:06 am
Hi Rue,
Betweenness is defined as the number of shortest paths between other nodes that passes through a node (node i in the equations). As there might be multiple shortest paths (same length and this is the shortest length), the ratio of g_jk(i)/g_jk ensure that double counting doesn’t occur (g_jk is the number of shortest paths between nodes j and k, and g_jk(i) is the number of these that passes through node i). In other words, if there are two paths that have the same length and this is the shortest length, then the nodes on both paths get a score of 1/2 assigned to them.
The g^w\alpha is the notation of shortest paths where the weights are incorporated and adjusted by the tuning parameter.
Best,
Tore
58. Rui | September 4, 2013 at 9:24 pm
Rui | September 4, 2013 at 9:24 pm
Hi Tore,
Thank you for the reply. I know what you mean. As you said in your example, g_jk(i)/g_jk is the score of 1/2, but this is not the betweenness of node i. You should add up g_jk(i)/g_jk for all pairs of j and k where j is not equal to k not equal to i.
My question is, since g_jk(i) and g_jk are the numbers of shortest paths, how you incorporated weights into g^w\alpha?
Best regards
Rui
59. Rui | September 5, 2013 at 8:55 am
Rui | September 5, 2013 at 8:55 am
Hi Tore,
I have got my answer. How silly I am…. “the weights are incorporated” doesn’t mean the weights are explicitly incorporated in g^w\alpha, but in the calculation of shortest paths. Thank you very much.
Best regards
Rui
60. Patrick S. Forscher | December 4, 2013 at 11:24 pm
Patrick S. Forscher | December 4, 2013 at 11:24 pm
Hi Tore,
Thanks very much for your work on weighted graphs and for your tnet package! I have two questions about your package.
I’ve been trying to use the tnet package to calculate centrality and centralization measures for a weighted, directed network. I noticed that igraph has implemented an option to calculate closeness for weighted, directed graphs, so I tried to duplicate my results from tnet in igraph to ensure that I was doing everything correctly. Below are my results using a toy network:
require(igraph)
require(tnet)
set.seed(1234)
m <- expand.grid(from = 1:4, to = 1:4)
m <- m[m$from != m$to, ]
m$weight <- sample(1:7, 12, replace = T)
igraph_g <- graph.data.frame(m)
tnet_g <- as.tnet(m)
closeness(igraph_g, mode = "in")
2 3 4 1
0.05882353 0.12500000 0.07692308 0.09090909
closeness(igraph_g, mode = "out")
2 3 4 1
0.12500000 0.06250000 0.06666667 0.10000000
closeness(igraph_g, mode = "total")
2 3 4 1
0.12500000 0.14285714 0.07692308 0.16666667
closeness_w(tnet_g, directed = T, alpha = 1)
node closeness n.closeness
[1,] 1 0.2721088 0.09070295
[2,] 2 0.2448980 0.08163265
[3,] 3 0.4130809 0.13769363
[4,] 4 0.4081633 0.13605442
Do you have any idea what might be causing the differing results between tnet and igraph?
My second question is a bit tougher. I would like to calculate the centralization of the closeness scores (and more broadly of the other centrality scores I obtain for my network). However, it's not obvious to me how to do this for a weighted network. Do you have any idea how I might calculate the centralization score for, say, the closeness centralities of a network?
61. Tore Opsahl | December 5, 2013 at 12:46 am
Tore Opsahl | December 5, 2013 at 12:46 am
Hi Patrick,
Thank you for using tnet! igraph is able to handle weights; however, the distance function in igraph expects weights that represent ‘costs’ instead of ‘strength’. In other words, the tie weight is considered the amount of energy needed to cross a tie. See Shortest Paths in Weighted Networks.
To your second question, I am afraid I cannot help. There are no good ways to my opinion to calculate centralization scores for weighted network. Even for binary networks, I do not agree with the centralization scores. They often assume that the node level scores grows with n-squared; however, this does not seem to be the case for networks. A better metric might be to report average degree etc.
Tore
62. Patrick S. Forscher | December 5, 2013 at 1:03 am
Patrick S. Forscher | December 5, 2013 at 1:03 am
Thank you very much for your prompt response! I am relieved to hear that the discrepancy between tnet and igraph is due to a simple scaling problem. However, I tried running the code that you provided and am still seeing a difference in the results from igraph and tnet. Here’s the matrix from tnet:
node closeness n.closeness
[1,] 1 0.2721088 0.09070295
[2,] 2 0.2448980 0.08163265
[3,] 3 0.4130809 0.13769363
[4,] 4 0.4081633 0.13605442
And here’s the vector from igraph:
2 3 4 1
0.3453947 0.1726974 0.1842105 0.2763158
Regarding centralizations for weighted networks, I’m a little disappointed to hear that there’s no good way to compute these. If I wanted to get a sense of, for example, how “cohesive” my network is based on the network weights, do you know of an analogue to the centralization that I could calculate? Or am I out of luck (at least until such measures are developed)?
63. Tore Opsahl | December 5, 2013 at 1:09 am
Tore Opsahl | December 5, 2013 at 1:09 am
Hi Patrick,
In an earlier version, I made a mistake. Please note that it is not a simple scaling issue between tnet and igraph in that you need to invert tie strengths for igraph.
> # Load packages > library(tnet) Loading required package: igraph Loading required package: survival Loading required package: splines tnet: Analysis of Weighted, Two-mode, and Longitudinal networks. Type ?tnet for help. > > # Create random network (you could also use the rg_w-function) > m <- expand.grid(from = 1:4, to = 1:4) > m <- m[m$from != m$to, ] > m$weight <- sample(1:7, 12, replace = T) > > # Make tnet object and calculate closeness > closeness_w(m) node closeness n.closeness [1,] 1 0.2193116 0.07310387 [2,] 2 0.3809524 0.12698413 [3,] 3 0.2825746 0.09419152 [4,] 4 0.3339518 0.11131725 > > # igraph > # Invert weights (transform into costs from strengths) > # Multiply weights by mean (just scaling, not really) > m$weight <- mean(m$weight)/m$weight > # Transform into igraph object > igraph_g <- graph.data.frame(m) > # Compute closeness > closeness(igraph_g, mode = "out") 2 3 4 1 0.3809524 0.2825746 0.3339518 0.2193116Note: igraph shows node 1 at the end of the list for some reason.
To test for cohesiveness, I would suggest density (only for binary networks) or the global clustering coefficient (binary and weighted networks; Clustering in Weighted Networks).
Hope this helps,
Tore
64. Patrick S. Forscher | December 5, 2013 at 1:12 am
Patrick S. Forscher | December 5, 2013 at 1:12 am
Aha, the revised code seems to do the trick, and based on a glance at the linked post, I think the global clustering coefficient is pretty close to what I want as a summary measure for my network.
Thanks again very much for your help!
65. Martin Warland | February 26, 2014 at 12:15 pm
Martin Warland | February 26, 2014 at 12:15 pm
Dear Tore,
Thank you for your terrific work! I find it very helpful. I want to compute node centrality in a weighted two mode network that takes into account weight of ties and number of ties.
Although you write a lot about two mode networks, I could unfortunately not find how I can apply “your “ node centrality measure to weighted two mode data.
Your response to the question of Mauricio (August 2010) was already very helpful in this regard. However when I run this with a sample data set, I get the response “In as.tnet(net, type = “weighted one-mode tnet”) : There were self-loops in the edgelist, these were removed”. I have used the following formula:
# Load tnet
> library(tnet)
>
> ## Read the undirected network
> weighted.net
> ## Run function
> degree_w(weighted.net, measure=c(“degree”,”output”,”alpha”), alpha=0.5)
node degree output alpha
[1,] 1 1 5 2.236068
[2,] 2 3 9 5.196152
[3,] 3 1 6 2.449490
[4,] 4 0 0 0.000000
[5,] 5 1 3 1.732051
[6,] 6 1 2 1.414214
How can I ensure that this runs with a weighted two mode tnet? From your manual (p.6) I understand that for a weighted (?) two mode network there need to be more than 4 and an unequal number of rows and columns. Therefore I have also tried it with 5 columns and more rows, yet I got the same response…
Does this function (https://toreopsahl.com/2011/08/08/degree-centrality-and-variation-in-tie-weights/) only apply to one mode data or can I use it for weighted two mode data as well? (for this I have tried to adjust (i.e. the type) it but still got some error messages)
I would very much appreciate your help.
Best wishes,
Martin
66. Tore Opsahl | February 26, 2014 at 2:47 pm
Tore Opsahl | February 26, 2014 at 2:47 pm
Hi Martin,
Thank you for your interest. I do believe that the issue is based on how the network is read. As weighted one-mode and two-mode networks both have a three column structure, it is not straight-forward to understand whether to treat the object as a one-mode or two-mode network. As one-mode networks are more common, as.tnet assumes three column objects to be one-mode networks. To specifically set an object to be read as a weighted two-mode network, run (assuming your data is loaded in an object called net):
net <- as.tnet(net, type='weighted two-mode tnet')
This should allow you to run all two-mode functions.
The degree measure post you are referring to is only for weighted one-mode networks (i.e., function name ends with _w). However, the measure could be applied to two mode data. While I don't recommend the following method for other functions, I believe it can be used for this case:
Then you should be able to run the metric.
Hope this helps,
Tore
67. Martin | February 26, 2014 at 4:14 pm
Martin | February 26, 2014 at 4:14 pm
Dear Tore
Thank you very much for impressively fast response. I am sorry, I still have remaining questions. Before I had already tried to include “net <- as.tnet(net, type='weighted two-mode tnet'” in the formula you are providing on this side: (https://toreopsahl.com/2011/08/08/degree-centrality-and-variation-in-tie-weights/, but I got error messages.
I understand that your proposed first formula is to avoid self-loops, isn’t it? In your second formula, you are suggesting to set it again as a one mode network. Does it mean that it is not possible to calculate node centrality (based on number of ties and weight of ties) in a weighted two mode network – without transferring it into one mode data?
Best wishes,
Martin
68. Tore Opsahl | February 27, 2014 at 1:53 am
Tore Opsahl | February 27, 2014 at 1:53 am
Martin,
I am unsure why you would get errors with the specification type=”weighted two-mode tnet”. If this is the case, please send me the code and data that you are using.
To your second point, indeed, the sample code shows how you can apply a one-mode metric to a two-mode network. However, this procedure only works for this measure as it is not focusing on the network, but more on the number and distribution of tie weights.
Do let me know if you have issues,
Tore
69. Leila | March 7, 2014 at 5:43 pm
Leila | March 7, 2014 at 5:43 pm
Dear Tore,
Thank you very much for your work and tnet, that I use a lot in my research papers. I have a question please. Is it possible to get easily in-closeness and out-closeness scores in a directed weighted network ? I know how to get in-degree and out degree but I can’t find the same distinction for closeness.
Thank you very much for your help.
Best Regards
LA
70. Tore Opsahl | March 8, 2014 at 3:43 pm
Tore Opsahl | March 8, 2014 at 3:43 pm
Hi Leila,
The closeness_w-function in tnet works by calculating the inverse of the sum of distances to other nodes. I would suggest that you reverse the ties. For example,
Hope this helps,
Tore
71. Leila | March 9, 2014 at 4:54 pm
Leila | March 9, 2014 at 4:54 pm
Thanks a lot Tore. I am sure It will work.
Kindest Regards
Leila
72. Li | March 10, 2014 at 7:19 am
Li | March 10, 2014 at 7:19 am
Hi Tore,
I am interested in using tnet to calculate weighted centrality for a one-mode network. I am having trouble reading the data into tnet. Would you please let me know where I got wrong?
“edge_PP 60.csv” is an edgelist with three columns, i, j, and weight without header.
el=read.csv(“edge_PP 60.csv”, header=FALSE)
el[,1]=as.character(el[,1])
el[,2]=as.character(el[,2])
el=as.matrix(el)
g=graph.edgelist(el[,1:2])
E(g)$weight=as.numeric(el[,3])
as.tnet(g)
Using above code, I got the following error message: “Error in if (NC == 2) net <- data.frame(tmp[, 1], tmp[, 2]) :
argument is of length zero"
Thank you for your suggestion!
73. Tore Opsahl | March 11, 2014 at 1:16 am
Tore Opsahl | March 11, 2014 at 1:16 am
Hi Li,
Great that you are using tnet! I would suggest that you do not load the network into igraph before loading it in tnet. If you transform the node identifiers into integers, you should be able to run as.tnet on the regular data.frame-object.
Best,
Tore
74. Li | April 3, 2014 at 4:58 pm
Li | April 3, 2014 at 4:58 pm
Hi Dr. Opsahl, this is Li again :-) I am wondering how we should deal with isolate in weighted network. Do we include them in the edgelist and assign the weight as 0? or we do not include them? If we do not include them, the size of the network will change. Does that matter?
Thanks for letting me know.
Best,
75. Tore Opsahl | April 6, 2014 at 8:48 pm
Tore Opsahl | April 6, 2014 at 8:48 pm
Hi Li,
Isolates should not be included in the edgelist as only ties are listed there. A jump in an edgelist assumes that isolates exists (e.g., see the code on the blog post for closeness in disconnection components where node K -an isolate- is assigned a score; https://toreopsahl.com/2010/03/20/closeness-centrality-in-networks-with-disconnected-components/).
For most metrics, a change in network size doesn’t matter (e.g., degree). However, if you calculate average degree, I would calculate degree and then sum the degree score and divide by the network size you have instead of simply writing mean(degree_w(net, measure=”degree”)[,”degree”]).
Hope this helps,
Tore
76. Pavlos Basaras | May 7, 2014 at 9:53 am
Pavlos Basaras | May 7, 2014 at 9:53 am
Hello Dr. Opsahl
i am using tnet to compute centrality measures (Closeness, Betweenness) in weighted directed networks using datasets from different sources (e.g http://snap.stanford.edu/data/links.html) and give probabilities as weights to the links in [0-1].
What i do in R is:
install.packages(“tnet”)
library(tnet)
directed.net <- read.table("C:\\Users\\Pavlos\\Desktop\\net.txt", sep="\t")
directed.net <- as.tnet(directed.net, type="weighted one-mode tnet")
sink("C:\\Users\\Pavlos\\Desktop\\bet.txt", append=FALSE, split=FALSE)
betweenness_w(directed.net, alpha=0.5)
sink("C:\\Users\\Pavlos\\Desktop\\clo.txt", append=FALSE, split=FALSE)
closeness_w(directed.net, alpha=0.5)
Everything seems to work perfect. First i would like to ask if i missed anything in the commands or if i did something wrong to this part. And also, i read to a previous post that weigths are accounted as costs, so a link of 0.4 will be prefered over a link of 0.9. Is that correct?
Many thanks for your time and work,
Pavlos
77. Tore Opsahl | May 7, 2014 at 11:22 pm
Tore Opsahl | May 7, 2014 at 11:22 pm
Hi Pavlos,
Great that you are applying the weighted metrics.
Your code seems fine. I would suggest that you take the output from the functions (e.g., closeness) and save it to an object, and then write that object to a file. But if the sink-function works for you, go ahead!
tnet assumes that a higher weight is better. This is in contrast to the shortest path functions of igraph, which assumes that people will invert the tie weights first themselves. From my own experience, we are not always so good at understanding the details of the functions before running them, so I have actually made tnet based on the common assumption of higher values = stronger ties = higher transmission. As such, a tie with a weight of 0.9 is preferred over a tie with 0.4.
Hope this clarifies!
Tore
78. Pavlos Basaras | May 8, 2014 at 7:32 am
Pavlos Basaras | May 8, 2014 at 7:32 am
Thank you very much for your prompt reply,
sink seems to work fine so i ‘ll leave it as it is for now.
I was thinking of inverting the weights so as to have the desired outcome but since you confirmed that tnet prefers higher values then i do not have to invert the weights and in both betweenness and closeness 0.9 will be prefered over 0.4, correct?
One more thing is that closeness is computed for the largest connected component, is there a way to compute it for the entire network topology?
Lastly in my work i am referencing the your website should i reference :
Opsahl, T., Agneessens, F., Skvoretz, J. (2010). Node centrality in weighted networks: Generalizing degree and shortest paths. Social Networks 32, 245-251.
instead of the webpage?
many thanks again,
Pavlos
79. Tore Opsahl | May 8, 2014 at 11:51 am
Tore Opsahl | May 8, 2014 at 11:51 am
Hi Pavlos,
Indeed. No need to invert tie weights in tnet.
To compute closeness for the full network, see https://toreopsahl.com/2010/03/20/closeness-centrality-in-networks-with-disconnected-components/ In other words, specify gconly=FALSE when running the function.
All the information on the site is in various papers, so you can just those. The relevant one for the information on a page is normally listed at the bottom of it.
Tore
80. Pavlos Basaras | May 8, 2014 at 12:04 pm
Pavlos Basaras | May 8, 2014 at 12:04 pm
Ok great,
thank you again,
Best,
Pavlos
81. Pavlos Basaras | June 3, 2014 at 6:37 am
Pavlos Basaras | June 3, 2014 at 6:37 am
Hello again,
i am using the version of closeness for the complete network topology and disconnected components as you suggested, however its seems that there is not enough memory to store the distance array for large networks. The networks i am testing are with nodes of size 83000, weighted and directed networks. I tested R in both 32-bit and 64-bit system and i also saw through other posts that it is a memory problem. How can we overcome this problem ?
Best,
Pavlos
82. Tore Opsahl | June 3, 2014 at 12:03 pm
Tore Opsahl | June 3, 2014 at 12:03 pm
Hi Pavlos,
There are two ways of dealing with running out of memory in R: buy more memory or make the code more memory-efficient. I had some time, and below is a suggestion for doing the latter. Note you might want to use JIT compiling to speed things up.
# To speed thing up, you might want to enable JIT compiling library(compiler) enableJIT(3) # Load tnet library(tnet) # Load sample network from blog post net <- cbind( i=c(1,1,2,2,2,3,3,3,4,4,4,5,5,6,6,7,9,10,10,11), j=c(2,3,1,3,5,1,2,4,3,6,7,2,6,4,5,4,10,9,11,10), w=c(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1)) # New function closeness_w2 <- function (net, directed = NULL, gconly = TRUE, alpha = 1) { if (is.null(attributes(net)$tnet)) net <- as.tnet(net, type = "weighted one-mode tnet") if (attributes(net)$tnet != "weighted one-mode tnet") stop("Network not loaded properly") net[, "w"] <- net[, "w"]^alpha if (is.null(directed)) { tmp <- symmetrise_w(net, method = "MAX") directed <- (nrow(tmp) != nrow(net) | sum(tmp[, "w"]) != sum(net[, "w"])) } # From distance_w-function g <- tnet_igraph(net, type = "weighted one-mode tnet", directed = directed) if (gconly) { stop("This code is only tested on gconly=FALSE") } else { gc <- as.integer(V(g)) } # Closeness scores out <- sapply(gc, function(a) { row <- as.numeric(igraph::shortest.paths(g, v=a, mode = "out", weights = igraph::get.edge.attribute(g,"tnetw"))) return(sum(1/row[row!=0])) }) out <- cbind(node = gc, closeness = out, n.closeness = out/(length(out) - 1)) return(out) } # JIT compiled closeness_w2c <- cmpfun(closeness_w2) # Scores with old function closeness_w(net, gconly=FALSE) # Scores with new function (regular and compiled) closeness_w2(net, gconly=FALSE) closeness_w2c(net, gconly=FALSE) # Disable JIT compiling enableJIT(0)Hope this helps!
Tore
83. Pavlos Basaras | June 4, 2014 at 12:58 pm
Pavlos Basaras | June 4, 2014 at 12:58 pm
Ok, it seems to work perfect, i used: closeness_w2c(net, directed=TRUE, gconly=FALSE)
for my results, thank you very much for your time and your help
Best,
Pavlos
84. Pavlos Basaras | June 18, 2014 at 8:54 am
Pavlos Basaras | June 18, 2014 at 8:54 am
Hello,
as you guided me in one of our previous discussions on how to use closeness on networks with disconnected components, is there a similar approach for betweenness in weighted and directed networks?
thanks you for your time,
Pavlos
85. Tore Opsahl | June 18, 2014 at 9:43 pm
Tore Opsahl | June 18, 2014 at 9:43 pm
Hi Pavlos,
Betweenness is not limited to the giant / main component; however, its distribution might be highly skewed and zero-inflated. Be sure to check before using the raw scores in frameworks that assumes a Gaussian distribution.
Best,
Tore
86. Pavlos Basaras | June 25, 2014 at 9:49 am
Pavlos Basaras | June 25, 2014 at 9:49 am
Hello,
actually the distribution i use is zipfian, ranging on skewness from 0.1 to 0.9. I get a lot zero values, (i assume this occurs because these nodes are not present in shorterst paths). Do you think there is a problem with zipfian ?
many thanks again,
Pavlos
87. Farnaz | November 17, 2014 at 8:59 am
Farnaz | November 17, 2014 at 8:59 am
hello Dr. opsahi
Thank you for your very good works.
i want to use this metric in the industrial engineering discipline, but i am a bit confused about the choose of alpha parameter, is that any procedure that i can use to determine the best value of alpha for my network? i mean that for example i consider degree is relatively more important for me, haw can i found which score i should choose?
many thanks.
Farnaz
88. Tore Opsahl | November 17, 2014 at 11:55 pm
Tore Opsahl | November 17, 2014 at 11:55 pm
Hi Farnaz,
There are two key values:
a) Alpha = 0 produces degree
b) Alpha = 1 produces strength
The closer alpha is set to 0, the more relevance is attached to degree over strength. See the discussion here: https://toreopsahl.com/tnet/weighted-networks/node-centrality/
Also, you might want to see comment #40 above. It outlines how to detect the most relevant alpha given an outcome / performance variable.
Hope this helps,
Tore
89. Nishtha | December 30, 2014 at 9:27 am
Nishtha | December 30, 2014 at 9:27 am
Good day!
Thank you for this awesome package. It is really useful. However, is there an upper limit to the number of nodes one can analyze using tnet? My data has 416 nodes, and every time I run I keep getting. Specifically, here is the code I have used:
set2008df =read.table(“test2008.txt”,header=TRUE) # read the data, this is in the form of i.j.w and is read as a dataframe.
set2008 closeness_w(set2008)
Error in unique(y[ind]) :
Value of SET_STRING_ELT() must be a ‘CHARSXP’ not a ‘bytecode’
Similarly, when I usee the degree_w(set2008), the following error:
*** caught segfault ***
address 0x0, cause ‘unknown’
Traceback:
1: cbind(1:max(net[, c(“i”, “j”)]), 0, 0, 0)
2: rbind(k.list, cbind(1:max(net[, c(“i”, “j”)]), 0, 0, 0))
3: degree_w(set2008)
Possible actions:
1: abort (with core dump, if enabled)
2: normal R exit
3: exit R without saving workspace
4: exit R saving workspace
Can you explain what is going on?
90. Tore Opsahl | January 1, 2015 at 11:06 pm
Tore Opsahl | January 1, 2015 at 11:06 pm
Hi Nishtha,
There seems to be some issues with your dataset. Are all the columns of the data.frame integer or numeric? Send me an email with the code and data if you have more issues.
Best,
Tore
91. Nishtha | February 7, 2015 at 3:48 am
Nishtha | February 7, 2015 at 3:48 am
Hi Tore:
The weight columns are numeric, the nodes are ids. I have also sent you the code and data. Let me know if you have more thoughts.
Thanks!
92. Zhitao Zhang | May 14, 2016 at 7:59 am
Zhitao Zhang | May 14, 2016 at 7:59 am
Hello Dr. Opsahi
Thank you for your good works. I want to use link betweenness to analyse some links’ intermediary in weighted networks.But you know the tnet only provides node betweenness’ function, could you give me some suggestions for solving this problem?
many thanks.
Zhitao Zhang
93. Tore Opsahl | May 17, 2016 at 3:16 am
Tore Opsahl | May 17, 2016 at 3:16 am
Hi Zhitao Zhang,
I do not know a specific implementation of link / edge betweenness for weighted networks. However, you can alter the tnet code to handle this case. Below is one example of such code.
Let me know if you are able to use it.
Tore
edge_betweenness_w <- function(net, directed = NULL, alpha = 1) { if (is.null(attributes(net)$tnet)) net <- as.tnet(net, type = "weighted one-mode tnet") if (attributes(net)$tnet != "weighted one-mode tnet") stop("Network not loaded properly") if (is.null(directed)) { tmp <- symmetrise_w(net, method = "MAX") directed <- (nrow(tmp) != nrow(net) | sum(tmp[, "w"]) != sum(net[, "w"])) } netTransformed <- net netTransformed[, "w"] <- (1/netTransformed[, "w"])^alpha g <- tnet_igraph(netTransformed, type = "weighted one-mode tnet", directed = directed) out <- data.frame(as_edgelist(g), betweenness = 0) colnames(out) <- c("i","j") out[, "betweenness"] <- igraph::edge_betweenness(graph = g, directed = directed) return(out) }94. Carmine | May 31, 2016 at 8:58 am
Carmine | May 31, 2016 at 8:58 am
Dear Tore,
thank you very much for excellent work. It would be extremely useful to use your measure of weighted centrality as I dispose of weighted interaction data. In particular my work focuses on inter-group interaction, and I would like to ask whether it would be possible to get a measure of weighted in-degree and out-degree centrality measure for inter-subgroup relationships.
Thank you in advance
Best
Carmine
95. Tore Opsahl | June 14, 2016 at 5:46 pm
Tore Opsahl | June 14, 2016 at 5:46 pm
Hi Carmine,
To get the out- and in-centrality measures, you need to first aggregate your network to the subgroup level (i.e., create a new network where the subgroups are the nodes) and then you can specify the type parameter as either “out” (default) or “in”.
Best,
Tore